Gene
KWMTBOMO02375
Pre Gene Modal
BGIBMGA013546
Annotation
PREDICTED:_phospholipid_scramblase_2-like_[Bombyx_mori]
Full name
Phospholipid scramblase
Location in the cell
Cytoplasmic Reliability : 1.4 Nuclear Reliability : 1.149
Sequence
CDS
ATGGCAACTTCGGCAATTTATGAAACTGCACCTCTGGGAATCCAACAGCTGATATCCTTAAGTAGCTTGAGGATCCAACAAAAGAAGACCCTTTGCGTCGGCAGCGAGTATACCGTGCTGAATGAAGACGGCGAAGAGATCCTTTACGGGAAGGAAGATGCCGCGGTCCTGAGTACGCTGTTCAGCGGGAAGCATCGTTCCTTCAACATGCACATTGTGGATAGTCAAGAAAATATGGTGATATTCCTTCGGCGGCCTCATACCTTGGGGCTGGACAAACTCGAGGTGAGAGTTGACAACGTCCTGGTCAGCATAGTGCGCCCTCAGCCGACATTCGTGAAGCCGGTGTTCAGCATCAACGACGCATCAGATAAACCGGTCCTTAGGGTCAAAGGCAAGCATTGTACCAGTGCAAATCACGAATATGAGGTGTTAGCGGTCAACAAAGAACGCATCGGTGGTATCAAGAAGCACGGAGCCGGCGTCGTGACCGAAATATGCACCAGTCATTATTACATCCAGTTCCCGGCCGACTTACATGTCGATTACAAAGCTGCTTTACTCGCCACTTGCTTCTTGATCGAATATATGTTCTTCGAAAAATGA
Protein
MATSAIYETAPLGIQQLISLSSLRIQQKKTLCVGSEYTVLNEDGEEILYGKEDAAVLSTLFSGKHRSFNMHIVDSQENMVIFLRRPHTLGLDKLEVRVDNVLVSIVRPQPTFVKPVFSINDASDKPVLRVKGKHCTSANHEYEVLAVNKERIGGIKKHGAGVVTEICTSHYYIQFPADLHVDYKAALLATCFLIEYMFFEK
Summary
Description
May mediate accelerated ATP-independent bidirectional transbilayer migration of phospholipids upon binding calcium ions that results in a loss of phospholipid asymmetry in the plasma membrane.
Cofactor
Ca(2+)
Similarity
Belongs to the phospholipid scramblase family.
Uniprot
H9JVI3
A0A194QAB6
A0A2A4J8J9
A0A2W1BJ50
A0A2H1WX23
A0A2H1VYI8
+ More
A0A212ERV1 A0A2M4CJ13 A0A2R7VYL5 A0A182M5Q3 A0A2A4J9L6 A0A2M3ZCD7 A0A182PN68 A0A2M4AMB1 A0A2M4AMH4 A0A2M4CRV1 A0A2M4AMK6 A0A2M4BT70 A0A2M4CT89 A0A182FF13 A0A084W2Y9 A0A182JKF2 A0A182H417 A0A1B6E985 A0A182KDU7 A0A1Y1LD75 W5JP83 A0A182WKA7 A0A182QML1 A0A182Y0X4 A7UTQ3 A0A182RN38 A0A182NM01 A0A182U4C8 E9GIA2 A0A182XIM0 A0A2C9GKK3 Q7PIS7 A0A182HM25 A7UTQ2 A0A182LJJ9 A0A023EP48 A0A182V3N8 A0A1Y1LB05 T1IB30 A0A0V0G640 A0A1S4FR97 Q16S90 B0WES2 A0A2W1BEL3 A0A1Q3FYP9 A0A2A4JBK1 A0A0P4VUN0 A0A1Q3G455 A0A1Q3G454 A0A069DSP2 A0A1B6J1R9 A0A1Q3G442 A0A1B6LTY2 A0A1B6LRE2 A0A1Q3G488 A0A224XTJ7 A0A0A9XAE0 A0A067QG92 A0A1W4WQ33 A0A1B6HDV9 T1K9R0 A0A0P6CLZ2 E0VXM0 A0A0P5SL04 A0A0P6CRD8 D6W8A6 A0A0P5S6U7 A0A2S2NZP4 T1K9Q9 N6TYF6 A0A1W4WF47 T1K9R3 A0A0P4WDV5 A0A0P5YAX0 A0A1L8DKT2 A0A1B6DDL0 A0A1B6BX50 A0A0P4WMS9 U5EVB8 A0A2J7R027 A0A0P6ESC2 A0A2H1W112 T1K9Q0 E9GIA0 E4WX33 U4TUV2 A0A2J7R040 A0A2J7R038 A0A336LIQ1 T1K9Q1 X1XPY5 A0A1B0CJF4 A2BEN3
A0A212ERV1 A0A2M4CJ13 A0A2R7VYL5 A0A182M5Q3 A0A2A4J9L6 A0A2M3ZCD7 A0A182PN68 A0A2M4AMB1 A0A2M4AMH4 A0A2M4CRV1 A0A2M4AMK6 A0A2M4BT70 A0A2M4CT89 A0A182FF13 A0A084W2Y9 A0A182JKF2 A0A182H417 A0A1B6E985 A0A182KDU7 A0A1Y1LD75 W5JP83 A0A182WKA7 A0A182QML1 A0A182Y0X4 A7UTQ3 A0A182RN38 A0A182NM01 A0A182U4C8 E9GIA2 A0A182XIM0 A0A2C9GKK3 Q7PIS7 A0A182HM25 A7UTQ2 A0A182LJJ9 A0A023EP48 A0A182V3N8 A0A1Y1LB05 T1IB30 A0A0V0G640 A0A1S4FR97 Q16S90 B0WES2 A0A2W1BEL3 A0A1Q3FYP9 A0A2A4JBK1 A0A0P4VUN0 A0A1Q3G455 A0A1Q3G454 A0A069DSP2 A0A1B6J1R9 A0A1Q3G442 A0A1B6LTY2 A0A1B6LRE2 A0A1Q3G488 A0A224XTJ7 A0A0A9XAE0 A0A067QG92 A0A1W4WQ33 A0A1B6HDV9 T1K9R0 A0A0P6CLZ2 E0VXM0 A0A0P5SL04 A0A0P6CRD8 D6W8A6 A0A0P5S6U7 A0A2S2NZP4 T1K9Q9 N6TYF6 A0A1W4WF47 T1K9R3 A0A0P4WDV5 A0A0P5YAX0 A0A1L8DKT2 A0A1B6DDL0 A0A1B6BX50 A0A0P4WMS9 U5EVB8 A0A2J7R027 A0A0P6ESC2 A0A2H1W112 T1K9Q0 E9GIA0 E4WX33 U4TUV2 A0A2J7R040 A0A2J7R038 A0A336LIQ1 T1K9Q1 X1XPY5 A0A1B0CJF4 A2BEN3
Pubmed
EMBL
BABH01040486
BABH01040487
BABH01040488
KQ459465
KPJ00356.1
NWSH01002372
+ More
PCG68447.1 KZ150157 PZC72760.1 ODYU01011703 SOQ57613.1 ODYU01005243 SOQ45910.1 AGBW02012923 OWR44205.1 GGFL01001174 MBW65352.1 KK854129 PTY11780.1 AXCM01002240 PCG68446.1 GGFM01005377 MBW26128.1 GGFK01008615 MBW41936.1 GGFK01008675 MBW41996.1 GGFL01003869 MBW68047.1 GGFK01008689 MBW42010.1 GGFJ01007145 MBW56286.1 GGFL01003870 MBW68048.1 ATLV01019732 KE525278 KFB44583.1 JXUM01108608 KQ565247 KXJ71181.1 GEDC01002799 JAS34499.1 GEZM01060886 JAV70838.1 ADMH02000550 ETN65911.1 AXCN02000941 AAAB01008960 EDO63675.1 GL732546 EFX80805.1 APCN01004326 APCN01004327 EAA44031.4 EDO63674.1 GAPW01002366 JAC11232.1 GEZM01060885 JAV70839.1 ACPB03021686 GECL01002646 JAP03478.1 CH477680 EAT37325.1 DS231911 EDS45835.1 PZC72761.1 GFDL01002387 JAV32658.1 NWSH01002038 PCG69341.1 GDKW01000186 JAI56409.1 GFDL01000464 JAV34581.1 GFDL01000462 JAV34583.1 GBGD01002138 JAC86751.1 GECU01014582 JAS93124.1 GFDL01000463 JAV34582.1 GEBQ01012851 JAT27126.1 GEBQ01013755 JAT26222.1 GFDL01000466 JAV34579.1 GFTR01005127 JAW11299.1 GBHO01025937 GBHO01025936 GDHC01020916 GDHC01012357 GDHC01004641 JAG17667.1 JAG17668.1 JAP97712.1 JAQ06272.1 JAQ13988.1 KK853500 KDR07158.1 GECU01034927 GECU01013232 JAS72779.1 JAS94474.1 CAEY01001892 GDIQ01089295 JAN05442.1 DS235833 EEB18126.1 GDIQ01091689 JAL60037.1 GDIQ01089294 JAN05443.1 KQ971307 EFA10920.2 GDIQ01091688 JAL60038.1 GGMR01009913 MBY22532.1 APGK01047306 KB741077 ENN74355.1 CAEY01001893 GDRN01078337 GDRN01078336 JAI62597.1 GDIP01060512 JAM43203.1 GFDF01007001 JAV07083.1 GEDC01013511 GEDC01011097 JAS23787.1 JAS26201.1 GEDC01031674 GEDC01011741 JAS05624.1 JAS25557.1 GDRN01078338 JAI62596.1 GANO01001947 JAB57924.1 NEVH01008299 PNF34192.1 GDIQ01071866 JAN22871.1 ODYU01005650 SOQ46717.1 EFX80807.1 FN653018 FN654894 CBY21925.1 CBY37071.1 KB631602 ERL84562.1 PNF34190.1 PNF34198.1 UFQS01000007 UFQT01000007 SSW97099.1 SSX17485.1 ABLF02035891 AJWK01014544 AJWK01014545 AJWK01014546 BX119902
PCG68447.1 KZ150157 PZC72760.1 ODYU01011703 SOQ57613.1 ODYU01005243 SOQ45910.1 AGBW02012923 OWR44205.1 GGFL01001174 MBW65352.1 KK854129 PTY11780.1 AXCM01002240 PCG68446.1 GGFM01005377 MBW26128.1 GGFK01008615 MBW41936.1 GGFK01008675 MBW41996.1 GGFL01003869 MBW68047.1 GGFK01008689 MBW42010.1 GGFJ01007145 MBW56286.1 GGFL01003870 MBW68048.1 ATLV01019732 KE525278 KFB44583.1 JXUM01108608 KQ565247 KXJ71181.1 GEDC01002799 JAS34499.1 GEZM01060886 JAV70838.1 ADMH02000550 ETN65911.1 AXCN02000941 AAAB01008960 EDO63675.1 GL732546 EFX80805.1 APCN01004326 APCN01004327 EAA44031.4 EDO63674.1 GAPW01002366 JAC11232.1 GEZM01060885 JAV70839.1 ACPB03021686 GECL01002646 JAP03478.1 CH477680 EAT37325.1 DS231911 EDS45835.1 PZC72761.1 GFDL01002387 JAV32658.1 NWSH01002038 PCG69341.1 GDKW01000186 JAI56409.1 GFDL01000464 JAV34581.1 GFDL01000462 JAV34583.1 GBGD01002138 JAC86751.1 GECU01014582 JAS93124.1 GFDL01000463 JAV34582.1 GEBQ01012851 JAT27126.1 GEBQ01013755 JAT26222.1 GFDL01000466 JAV34579.1 GFTR01005127 JAW11299.1 GBHO01025937 GBHO01025936 GDHC01020916 GDHC01012357 GDHC01004641 JAG17667.1 JAG17668.1 JAP97712.1 JAQ06272.1 JAQ13988.1 KK853500 KDR07158.1 GECU01034927 GECU01013232 JAS72779.1 JAS94474.1 CAEY01001892 GDIQ01089295 JAN05442.1 DS235833 EEB18126.1 GDIQ01091689 JAL60037.1 GDIQ01089294 JAN05443.1 KQ971307 EFA10920.2 GDIQ01091688 JAL60038.1 GGMR01009913 MBY22532.1 APGK01047306 KB741077 ENN74355.1 CAEY01001893 GDRN01078337 GDRN01078336 JAI62597.1 GDIP01060512 JAM43203.1 GFDF01007001 JAV07083.1 GEDC01013511 GEDC01011097 JAS23787.1 JAS26201.1 GEDC01031674 GEDC01011741 JAS05624.1 JAS25557.1 GDRN01078338 JAI62596.1 GANO01001947 JAB57924.1 NEVH01008299 PNF34192.1 GDIQ01071866 JAN22871.1 ODYU01005650 SOQ46717.1 EFX80807.1 FN653018 FN654894 CBY21925.1 CBY37071.1 KB631602 ERL84562.1 PNF34190.1 PNF34198.1 UFQS01000007 UFQT01000007 SSW97099.1 SSX17485.1 ABLF02035891 AJWK01014544 AJWK01014545 AJWK01014546 BX119902
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000075883
UP000075885
+ More
UP000069272 UP000030765 UP000075880 UP000069940 UP000249989 UP000075881 UP000000673 UP000075920 UP000075886 UP000076408 UP000007062 UP000075900 UP000075884 UP000075902 UP000000305 UP000076407 UP000075840 UP000075882 UP000075903 UP000015103 UP000008820 UP000002320 UP000027135 UP000192223 UP000015104 UP000009046 UP000007266 UP000019118 UP000235965 UP000030742 UP000007819 UP000092461 UP000000437
UP000069272 UP000030765 UP000075880 UP000069940 UP000249989 UP000075881 UP000000673 UP000075920 UP000075886 UP000076408 UP000007062 UP000075900 UP000075884 UP000075902 UP000000305 UP000076407 UP000075840 UP000075882 UP000075903 UP000015103 UP000008820 UP000002320 UP000027135 UP000192223 UP000015104 UP000009046 UP000007266 UP000019118 UP000235965 UP000030742 UP000007819 UP000092461 UP000000437
PRIDE
SUPFAM
SSF54518
SSF54518
Gene 3D
ProteinModelPortal
H9JVI3
A0A194QAB6
A0A2A4J8J9
A0A2W1BJ50
A0A2H1WX23
A0A2H1VYI8
+ More
A0A212ERV1 A0A2M4CJ13 A0A2R7VYL5 A0A182M5Q3 A0A2A4J9L6 A0A2M3ZCD7 A0A182PN68 A0A2M4AMB1 A0A2M4AMH4 A0A2M4CRV1 A0A2M4AMK6 A0A2M4BT70 A0A2M4CT89 A0A182FF13 A0A084W2Y9 A0A182JKF2 A0A182H417 A0A1B6E985 A0A182KDU7 A0A1Y1LD75 W5JP83 A0A182WKA7 A0A182QML1 A0A182Y0X4 A7UTQ3 A0A182RN38 A0A182NM01 A0A182U4C8 E9GIA2 A0A182XIM0 A0A2C9GKK3 Q7PIS7 A0A182HM25 A7UTQ2 A0A182LJJ9 A0A023EP48 A0A182V3N8 A0A1Y1LB05 T1IB30 A0A0V0G640 A0A1S4FR97 Q16S90 B0WES2 A0A2W1BEL3 A0A1Q3FYP9 A0A2A4JBK1 A0A0P4VUN0 A0A1Q3G455 A0A1Q3G454 A0A069DSP2 A0A1B6J1R9 A0A1Q3G442 A0A1B6LTY2 A0A1B6LRE2 A0A1Q3G488 A0A224XTJ7 A0A0A9XAE0 A0A067QG92 A0A1W4WQ33 A0A1B6HDV9 T1K9R0 A0A0P6CLZ2 E0VXM0 A0A0P5SL04 A0A0P6CRD8 D6W8A6 A0A0P5S6U7 A0A2S2NZP4 T1K9Q9 N6TYF6 A0A1W4WF47 T1K9R3 A0A0P4WDV5 A0A0P5YAX0 A0A1L8DKT2 A0A1B6DDL0 A0A1B6BX50 A0A0P4WMS9 U5EVB8 A0A2J7R027 A0A0P6ESC2 A0A2H1W112 T1K9Q0 E9GIA0 E4WX33 U4TUV2 A0A2J7R040 A0A2J7R038 A0A336LIQ1 T1K9Q1 X1XPY5 A0A1B0CJF4 A2BEN3
A0A212ERV1 A0A2M4CJ13 A0A2R7VYL5 A0A182M5Q3 A0A2A4J9L6 A0A2M3ZCD7 A0A182PN68 A0A2M4AMB1 A0A2M4AMH4 A0A2M4CRV1 A0A2M4AMK6 A0A2M4BT70 A0A2M4CT89 A0A182FF13 A0A084W2Y9 A0A182JKF2 A0A182H417 A0A1B6E985 A0A182KDU7 A0A1Y1LD75 W5JP83 A0A182WKA7 A0A182QML1 A0A182Y0X4 A7UTQ3 A0A182RN38 A0A182NM01 A0A182U4C8 E9GIA2 A0A182XIM0 A0A2C9GKK3 Q7PIS7 A0A182HM25 A7UTQ2 A0A182LJJ9 A0A023EP48 A0A182V3N8 A0A1Y1LB05 T1IB30 A0A0V0G640 A0A1S4FR97 Q16S90 B0WES2 A0A2W1BEL3 A0A1Q3FYP9 A0A2A4JBK1 A0A0P4VUN0 A0A1Q3G455 A0A1Q3G454 A0A069DSP2 A0A1B6J1R9 A0A1Q3G442 A0A1B6LTY2 A0A1B6LRE2 A0A1Q3G488 A0A224XTJ7 A0A0A9XAE0 A0A067QG92 A0A1W4WQ33 A0A1B6HDV9 T1K9R0 A0A0P6CLZ2 E0VXM0 A0A0P5SL04 A0A0P6CRD8 D6W8A6 A0A0P5S6U7 A0A2S2NZP4 T1K9Q9 N6TYF6 A0A1W4WF47 T1K9R3 A0A0P4WDV5 A0A0P5YAX0 A0A1L8DKT2 A0A1B6DDL0 A0A1B6BX50 A0A0P4WMS9 U5EVB8 A0A2J7R027 A0A0P6ESC2 A0A2H1W112 T1K9Q0 E9GIA0 E4WX33 U4TUV2 A0A2J7R040 A0A2J7R038 A0A336LIQ1 T1K9Q1 X1XPY5 A0A1B0CJF4 A2BEN3
Ontologies
PANTHER
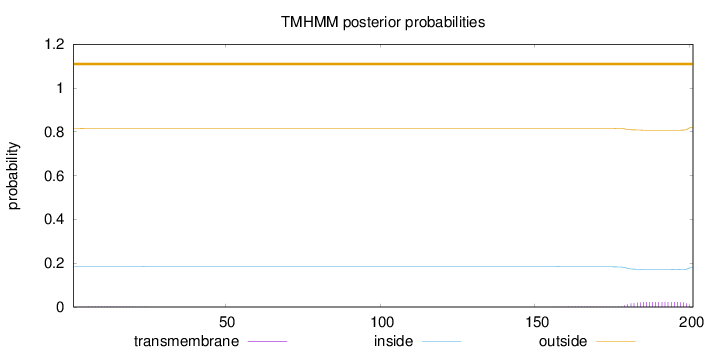
Topology
Length:
201
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.47581
Exp number, first 60 AAs:
0.0248
Total prob of N-in:
0.18376
outside
1 - 201
Population Genetic Test Statistics
Pi
131.402053
Theta
151.818139
Tajima's D
0.4756
CLR
1.860855
CSRT
0.500824958752062
Interpretation
Uncertain
