Gene
KWMTBOMO02354 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014417
Annotation
PREDICTED:_very_long-chain_specific_acyl-CoA_dehydrogenase?_mitochondrial_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.356
Sequence
CDS
ATGAAAGGTGCCAAGCTGGTCCTGTGTGGCGGTCGCTGCCTCGTCCCCAAAACGACTGCGGTGGTTCCGCTTTGCAGCGGAGCTCGATCTGCCGCATCAGAGGCGCAAGCTCGAAACGGACCGAGACAGAGTGCCTCTTTCACTCTGAATCTTTTCCGTGGCCGATTTGAACCTACACAGGTGTTTCCTTTTCCGGAACCACTTTCCGATGACCAACGTCAAATGCTACAAGAGCTCGTGCCGCCAGTCGAGAAATTCTTTCAAGAAGTAAATGACCCCGCTAAGAATGATGAGAATTCAAAGATTGAGGAGGGTACCGTCGCCGGACTGTGGGATCTCGGCGCGTTCGGGCTGCAGGTGCCGGGGGAGCTCGGCGGCCTGGGGCTGTCCAACACGCAGTACGCGCGGCTCGTCGAGGTGGTCGGGGCCCACGACCTCGGCGTCGGCATCACGCTCGGGGCGCATCAGTCCATCGGCTTCAAGGGAATCTTGCTGTTCGGCAACCCGCAGCAGAAAGAGCATTACCTGCCGCGCGTCACTTCCGGCGAGTATGCCGCCTTCTGCCTCACCGAACCTTCTTCCGGTTCAGACGCCGGTTCTATCAAGACTCGAGCCGTGCTGGCGCCCGACGGCAAACATTTCATCCTAAATGGTTCGAAGATCTGGATCAGCAACGGCGGTATCGCAGAGATTATGACGGTTTTCGCACAAACTCCTATCGAGAAGGACGGAAAGACCGTTGATAAGGTGACGGCGTTCATCGTGGAGAGGTCGTTCGGCGGCGTGTCGTCCGGCCCGCCCGAGAACAAGATGGGCATCAAGTGCTCCAACACCACGGAGGTGTACTACGAGGACGTGCGCGTGCCCGTCGAGAACGTGCTGGGCGGCGTCGGGAACGGGTTCAAGGTCGCCATGAACATCCTCAACAACGGGCGCTTCGGCATGGCGGCCGCGCTGGCCGGCACGCAGCGCGCCGCCATCGCGCAGGCCGCGCAGCACGCCGCCACGCGCGTGCAGTTCGGGAAGCACCTGGCCGAGTTCGGCGCCGTGCAGGAGAAGCTGGCCCGCATGGCCATGCTGCAGTACGCCACCGAGGCGCTCGCCTACATGGTCTCCGGCAACATGGACTCCGGCGCGCAGGACTACCACCTGGAGGCCGCCATCTCGAAGGTTTTCGCTTCGGACAGCGCATGGACAGTCGTCGATGAAGCCATCCAGATCCTCGGCGGGATGGGCTACATGAAGGCGACGGGGCTGGAGCGCGTGCTGCGCGATCTCCGCATCTTCCGCATCTTCGAAGGCACCAACGACATCCTGCGTCTGTTCGTGGCGCTCACGGGAATCCAGTTCGCCGGATCTCACCTCCAGGAGCTCCAGCGAGCCTTCAAGAACCCGACCGCACATCTCGGGCTCATATTCTCGGAGGCCGGGCGGCGCGCCACGGCCGCCGTGGGGCTGGCGCGCGGGGCGGACCTGAGCTCGCTGGTGGCGCCCGAGCTGCGGGGCGCGGCGGCCGAGCTGGGCAAGCTGGTGGGCGAGCACGGGCGCGCGGTGGAGGCGGTGCTGCTGAGGCACGGGCGCGGCGTGGTGGAGCAGCAGTGCGTGCTCAACCGGCTGGCGGGCGCGGCCGTGGACGCGTACACGTCGGCCGCCGTGCTGTCCCGCGCCTCCCGCGCCGCGCGCCGCTCGCTGCCCGGCGCCGCGCACGAGCTGCGCCTGGCCGCCGCGTGGGCCGAGGAGGCGCTGGGCCGCATCCCGGCGCAGCTGGCCGCCACCCGGGACGCGCGCTCCCAGGCGCACTTCCAGCGGCTCGGCGCGCTGGGCCGCGACGTGGCCGACCAGGGCGGCCAGCTGTGTCCCAACCCGCTCTACTTGTGA
Protein
MKGAKLVLCGGRCLVPKTTAVVPLCSGARSAASEAQARNGPRQSASFTLNLFRGRFEPTQVFPFPEPLSDDQRQMLQELVPPVEKFFQEVNDPAKNDENSKIEEGTVAGLWDLGAFGLQVPGELGGLGLSNTQYARLVEVVGAHDLGVGITLGAHQSIGFKGILLFGNPQQKEHYLPRVTSGEYAAFCLTEPSSGSDAGSIKTRAVLAPDGKHFILNGSKIWISNGGIAEIMTVFAQTPIEKDGKTVDKVTAFIVERSFGGVSSGPPENKMGIKCSNTTEVYYEDVRVPVENVLGGVGNGFKVAMNILNNGRFGMAAALAGTQRAAIAQAAQHAATRVQFGKHLAEFGAVQEKLARMAMLQYATEALAYMVSGNMDSGAQDYHLEAAISKVFASDSAWTVVDEAIQILGGMGYMKATGLERVLRDLRIFRIFEGTNDILRLFVALTGIQFAGSHLQELQRAFKNPTAHLGLIFSEAGRRATAAVGLARGADLSSLVAPELRGAAAELGKLVGEHGRAVEAVLLRHGRGVVEQQCVLNRLAGAAVDAYTSAAVLSRASRAARRSLPGAAHELRLAAAWAEEALGRIPAQLAATRDARSQAHFQRLGALGRDVADQGGQLCPNPLYL
Summary
Cofactor
FAD
Similarity
Belongs to the acyl-CoA dehydrogenase family.
Uniprot
A0A212FAQ9
A0A2A4JJJ5
A0A2H1W8W9
A0A068FJE7
A0A194Q5T9
S4PA42
+ More
A0A2H8TU38 J9JSE3 A0A0A9YWD2 A0A146KS87 A0A2R7WAY7 A0A2J7QAH9 A0A067R0D8 D6W7D3 A0A2W1BJG1 A0A0V0G6X1 A0A2S2NRD6 A0A023F5E9 A0A224X9J1 A0A2S2R506 A0A1B6KGK5 A0A0P4VJV3 A0A1B6DU68 A0A0L7L6X0 T1ICS4 A0A1B6I437 N6T8U6 A0A1B6G2Y9 U4UJI2 A0A0K8SGZ5 K7J7J5 A0A0J7KZ01 A0A0L7QS87 A0A1W4XHL4 A0A154PP36 A0A0C9REM8 A0A0C9RAV1 E2AEK8 E2BYV9 A0A151WJV4 A0A195EX02 A0A151IWW1 A0A195BTK6 A0A158NNW9 A0A084VMQ9 A0A151I956 F4WV82 A0A3L8DKY6 Q17BX4 E9J061 A0A1L8DZ29 A0A1Q3FHM4 A0A1Q3FI21 A0A1I8PSL0 A0A0P4W1G9 A0A182NB72 A0A1Q3FGK6 A0A023EVD3 A0A182JUD3 A0A1Y1KZ03 A0A182RA72 A0A182J7F3 A0A1B0DB09 A0A182X4Z8 A0A1B0GKL3 A0A182Y8I9 A0A182GZ27 A0A0L0CK58 A0A2M4A3R3 A0A1A9ZX86 W5J382 A0A182QF10 A0A182KWM7 A0A1B0C593 A0A182SNB6 A0A182VFM0 A0A182PUH8 A0A2M4A486 D3TR49 A0A1S4HD25 A0A182TFN3 A0A1I8JU27 V4BBD4 B0XAD1 A0A182GT47 A0A182MJ79 U5EKH4 A0A1A9Y1X5 A0A2M4A4K7 A0A336M2Q0 A0A226EQX8 A0A2M3Z1C8 A0A182VTL8 A0A1Y9G9M4 A0A2G8KXD8 A0A1A9UIN8 A0A026WQW2 A0A1W4V026 A0A2M4BH60 A0A1D2MYK9
A0A2H8TU38 J9JSE3 A0A0A9YWD2 A0A146KS87 A0A2R7WAY7 A0A2J7QAH9 A0A067R0D8 D6W7D3 A0A2W1BJG1 A0A0V0G6X1 A0A2S2NRD6 A0A023F5E9 A0A224X9J1 A0A2S2R506 A0A1B6KGK5 A0A0P4VJV3 A0A1B6DU68 A0A0L7L6X0 T1ICS4 A0A1B6I437 N6T8U6 A0A1B6G2Y9 U4UJI2 A0A0K8SGZ5 K7J7J5 A0A0J7KZ01 A0A0L7QS87 A0A1W4XHL4 A0A154PP36 A0A0C9REM8 A0A0C9RAV1 E2AEK8 E2BYV9 A0A151WJV4 A0A195EX02 A0A151IWW1 A0A195BTK6 A0A158NNW9 A0A084VMQ9 A0A151I956 F4WV82 A0A3L8DKY6 Q17BX4 E9J061 A0A1L8DZ29 A0A1Q3FHM4 A0A1Q3FI21 A0A1I8PSL0 A0A0P4W1G9 A0A182NB72 A0A1Q3FGK6 A0A023EVD3 A0A182JUD3 A0A1Y1KZ03 A0A182RA72 A0A182J7F3 A0A1B0DB09 A0A182X4Z8 A0A1B0GKL3 A0A182Y8I9 A0A182GZ27 A0A0L0CK58 A0A2M4A3R3 A0A1A9ZX86 W5J382 A0A182QF10 A0A182KWM7 A0A1B0C593 A0A182SNB6 A0A182VFM0 A0A182PUH8 A0A2M4A486 D3TR49 A0A1S4HD25 A0A182TFN3 A0A1I8JU27 V4BBD4 B0XAD1 A0A182GT47 A0A182MJ79 U5EKH4 A0A1A9Y1X5 A0A2M4A4K7 A0A336M2Q0 A0A226EQX8 A0A2M3Z1C8 A0A182VTL8 A0A1Y9G9M4 A0A2G8KXD8 A0A1A9UIN8 A0A026WQW2 A0A1W4V026 A0A2M4BH60 A0A1D2MYK9
Pubmed
22118469
26385554
26354079
23622113
25401762
26823975
+ More
24845553 18362917 19820115 28756777 25474469 27129103 26227816 23537049 20075255 20798317 21347285 24438588 21719571 30249741 17510324 21282665 24945155 28004739 25244985 26483478 26108605 20920257 23761445 20966253 20353571 12364791 23254933 29023486 24508170 27289101
24845553 18362917 19820115 28756777 25474469 27129103 26227816 23537049 20075255 20798317 21347285 24438588 21719571 30249741 17510324 21282665 24945155 28004739 25244985 26483478 26108605 20920257 23761445 20966253 20353571 12364791 23254933 29023486 24508170 27289101
EMBL
AGBW02009439
OWR50809.1
NWSH01001272
PCG71898.1
ODYU01007043
SOQ49463.1
+ More
KJ622082 AID66672.1 KQ459465 KPJ00370.1 GAIX01003419 JAA89141.1 GFXV01004893 MBW16698.1 ABLF02032168 GBHO01036805 GBHO01036801 GBHO01036797 GBHO01006282 JAG06799.1 JAG06803.1 JAG06807.1 JAG37322.1 GDHC01020537 GDHC01014106 JAP98091.1 JAQ04523.1 KK854536 PTY16678.1 NEVH01016330 PNF25569.1 KK852804 KDR16191.1 KQ971307 EFA11043.1 KZ150107 PZC73397.1 GECL01002266 JAP03858.1 GGMR01006953 MBY19572.1 GBBI01002348 JAC16364.1 GFTR01007420 JAW09006.1 GGMS01015845 MBY85048.1 GEBQ01029404 JAT10573.1 GDKW01001288 JAI55307.1 GEDC01028317 GEDC01008076 JAS08981.1 JAS29222.1 JTDY01002612 KOB71094.1 ACPB03002965 GECU01026041 JAS81665.1 APGK01047094 KB741077 ENN74123.1 GECZ01012959 JAS56810.1 KB632384 ERL94254.1 GBRD01013422 JAG52404.1 LBMM01001769 KMQ95777.1 KQ414766 KOC61404.1 KQ435007 KZC13622.1 GBYB01005441 JAG75208.1 GBYB01005440 JAG75207.1 GL438870 EFN68131.1 GL451531 EFN79131.1 KQ983031 KYQ48142.1 KQ981953 KYN32427.1 KQ980843 KYN12315.1 KQ976417 KYM89759.1 ADTU01021860 ATLV01014619 KE524975 KFB39253.1 KQ978303 KYM95338.1 GL888384 EGI61848.1 QOIP01000006 RLU21100.1 CH477314 EAT43846.1 GL767291 EFZ13851.1 GFDF01002353 JAV11731.1 GFDL01007988 JAV27057.1 GFDL01007829 JAV27216.1 GDRN01080252 JAI62261.1 GFDL01008355 JAV26690.1 GAPW01000505 JAC13093.1 GEZM01075386 JAV64107.1 AJVK01000593 AJWK01029092 AJWK01029093 JXUM01098615 KQ564435 KXJ72282.1 JRES01000301 KNC32597.1 GGFK01002060 MBW35381.1 ADMH02002211 ETN57783.1 AXCN02000637 JXJN01025895 GGFK01002292 MBW35613.1 CCAG010011281 EZ423901 ADD20177.1 AAAB01008944 APCN01000643 KB199651 ESP04846.1 DS232578 EDS43628.1 JXUM01086210 JXUM01086211 JXUM01086212 KQ563552 KXJ73666.1 AXCM01004246 GANO01001812 JAB58059.1 GGFK01002423 MBW35744.1 UFQS01000326 UFQT01000326 SSX02912.1 SSX23279.1 LNIX01000002 OXA60032.1 GGFM01001555 MBW22306.1 MRZV01000318 PIK52677.1 KK107128 EZA58348.1 GGFJ01003231 MBW52372.1 LJIJ01000387 ODM98031.1
KJ622082 AID66672.1 KQ459465 KPJ00370.1 GAIX01003419 JAA89141.1 GFXV01004893 MBW16698.1 ABLF02032168 GBHO01036805 GBHO01036801 GBHO01036797 GBHO01006282 JAG06799.1 JAG06803.1 JAG06807.1 JAG37322.1 GDHC01020537 GDHC01014106 JAP98091.1 JAQ04523.1 KK854536 PTY16678.1 NEVH01016330 PNF25569.1 KK852804 KDR16191.1 KQ971307 EFA11043.1 KZ150107 PZC73397.1 GECL01002266 JAP03858.1 GGMR01006953 MBY19572.1 GBBI01002348 JAC16364.1 GFTR01007420 JAW09006.1 GGMS01015845 MBY85048.1 GEBQ01029404 JAT10573.1 GDKW01001288 JAI55307.1 GEDC01028317 GEDC01008076 JAS08981.1 JAS29222.1 JTDY01002612 KOB71094.1 ACPB03002965 GECU01026041 JAS81665.1 APGK01047094 KB741077 ENN74123.1 GECZ01012959 JAS56810.1 KB632384 ERL94254.1 GBRD01013422 JAG52404.1 LBMM01001769 KMQ95777.1 KQ414766 KOC61404.1 KQ435007 KZC13622.1 GBYB01005441 JAG75208.1 GBYB01005440 JAG75207.1 GL438870 EFN68131.1 GL451531 EFN79131.1 KQ983031 KYQ48142.1 KQ981953 KYN32427.1 KQ980843 KYN12315.1 KQ976417 KYM89759.1 ADTU01021860 ATLV01014619 KE524975 KFB39253.1 KQ978303 KYM95338.1 GL888384 EGI61848.1 QOIP01000006 RLU21100.1 CH477314 EAT43846.1 GL767291 EFZ13851.1 GFDF01002353 JAV11731.1 GFDL01007988 JAV27057.1 GFDL01007829 JAV27216.1 GDRN01080252 JAI62261.1 GFDL01008355 JAV26690.1 GAPW01000505 JAC13093.1 GEZM01075386 JAV64107.1 AJVK01000593 AJWK01029092 AJWK01029093 JXUM01098615 KQ564435 KXJ72282.1 JRES01000301 KNC32597.1 GGFK01002060 MBW35381.1 ADMH02002211 ETN57783.1 AXCN02000637 JXJN01025895 GGFK01002292 MBW35613.1 CCAG010011281 EZ423901 ADD20177.1 AAAB01008944 APCN01000643 KB199651 ESP04846.1 DS232578 EDS43628.1 JXUM01086210 JXUM01086211 JXUM01086212 KQ563552 KXJ73666.1 AXCM01004246 GANO01001812 JAB58059.1 GGFK01002423 MBW35744.1 UFQS01000326 UFQT01000326 SSX02912.1 SSX23279.1 LNIX01000002 OXA60032.1 GGFM01001555 MBW22306.1 MRZV01000318 PIK52677.1 KK107128 EZA58348.1 GGFJ01003231 MBW52372.1 LJIJ01000387 ODM98031.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000007819
UP000235965
UP000027135
+ More
UP000007266 UP000037510 UP000015103 UP000019118 UP000030742 UP000002358 UP000036403 UP000053825 UP000192223 UP000076502 UP000000311 UP000008237 UP000075809 UP000078541 UP000078492 UP000078540 UP000005205 UP000030765 UP000078542 UP000007755 UP000279307 UP000008820 UP000095300 UP000075884 UP000075881 UP000075900 UP000075880 UP000092462 UP000076407 UP000092461 UP000076408 UP000069940 UP000249989 UP000037069 UP000092445 UP000000673 UP000075886 UP000075882 UP000092460 UP000075901 UP000075903 UP000075885 UP000092444 UP000075902 UP000075840 UP000030746 UP000002320 UP000075883 UP000092443 UP000198287 UP000075920 UP000069272 UP000230750 UP000078200 UP000053097 UP000192221 UP000094527
UP000007266 UP000037510 UP000015103 UP000019118 UP000030742 UP000002358 UP000036403 UP000053825 UP000192223 UP000076502 UP000000311 UP000008237 UP000075809 UP000078541 UP000078492 UP000078540 UP000005205 UP000030765 UP000078542 UP000007755 UP000279307 UP000008820 UP000095300 UP000075884 UP000075881 UP000075900 UP000075880 UP000092462 UP000076407 UP000092461 UP000076408 UP000069940 UP000249989 UP000037069 UP000092445 UP000000673 UP000075886 UP000075882 UP000092460 UP000075901 UP000075903 UP000075885 UP000092444 UP000075902 UP000075840 UP000030746 UP000002320 UP000075883 UP000092443 UP000198287 UP000075920 UP000069272 UP000230750 UP000078200 UP000053097 UP000192221 UP000094527
Pfam
Interpro
IPR036250
AcylCo_DH-like_C
+ More
IPR037069 AcylCoA_DH/ox_N_sf
IPR006089 Acyl-CoA_DH_CS
IPR013786 AcylCoA_DH/ox_N
IPR009075 AcylCo_DH/oxidase_C
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR006578 MADF-dom
IPR011990 TPR-like_helical_dom_sf
IPR033490 LRP130
IPR002885 Pentatricopeptide_repeat
IPR033443 PPR_long
IPR029001 ITPase-like_fam
IPR003697 Maf-like
IPR037069 AcylCoA_DH/ox_N_sf
IPR006089 Acyl-CoA_DH_CS
IPR013786 AcylCoA_DH/ox_N
IPR009075 AcylCo_DH/oxidase_C
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR006578 MADF-dom
IPR011990 TPR-like_helical_dom_sf
IPR033490 LRP130
IPR002885 Pentatricopeptide_repeat
IPR033443 PPR_long
IPR029001 ITPase-like_fam
IPR003697 Maf-like
Gene 3D
CDD
ProteinModelPortal
A0A212FAQ9
A0A2A4JJJ5
A0A2H1W8W9
A0A068FJE7
A0A194Q5T9
S4PA42
+ More
A0A2H8TU38 J9JSE3 A0A0A9YWD2 A0A146KS87 A0A2R7WAY7 A0A2J7QAH9 A0A067R0D8 D6W7D3 A0A2W1BJG1 A0A0V0G6X1 A0A2S2NRD6 A0A023F5E9 A0A224X9J1 A0A2S2R506 A0A1B6KGK5 A0A0P4VJV3 A0A1B6DU68 A0A0L7L6X0 T1ICS4 A0A1B6I437 N6T8U6 A0A1B6G2Y9 U4UJI2 A0A0K8SGZ5 K7J7J5 A0A0J7KZ01 A0A0L7QS87 A0A1W4XHL4 A0A154PP36 A0A0C9REM8 A0A0C9RAV1 E2AEK8 E2BYV9 A0A151WJV4 A0A195EX02 A0A151IWW1 A0A195BTK6 A0A158NNW9 A0A084VMQ9 A0A151I956 F4WV82 A0A3L8DKY6 Q17BX4 E9J061 A0A1L8DZ29 A0A1Q3FHM4 A0A1Q3FI21 A0A1I8PSL0 A0A0P4W1G9 A0A182NB72 A0A1Q3FGK6 A0A023EVD3 A0A182JUD3 A0A1Y1KZ03 A0A182RA72 A0A182J7F3 A0A1B0DB09 A0A182X4Z8 A0A1B0GKL3 A0A182Y8I9 A0A182GZ27 A0A0L0CK58 A0A2M4A3R3 A0A1A9ZX86 W5J382 A0A182QF10 A0A182KWM7 A0A1B0C593 A0A182SNB6 A0A182VFM0 A0A182PUH8 A0A2M4A486 D3TR49 A0A1S4HD25 A0A182TFN3 A0A1I8JU27 V4BBD4 B0XAD1 A0A182GT47 A0A182MJ79 U5EKH4 A0A1A9Y1X5 A0A2M4A4K7 A0A336M2Q0 A0A226EQX8 A0A2M3Z1C8 A0A182VTL8 A0A1Y9G9M4 A0A2G8KXD8 A0A1A9UIN8 A0A026WQW2 A0A1W4V026 A0A2M4BH60 A0A1D2MYK9
A0A2H8TU38 J9JSE3 A0A0A9YWD2 A0A146KS87 A0A2R7WAY7 A0A2J7QAH9 A0A067R0D8 D6W7D3 A0A2W1BJG1 A0A0V0G6X1 A0A2S2NRD6 A0A023F5E9 A0A224X9J1 A0A2S2R506 A0A1B6KGK5 A0A0P4VJV3 A0A1B6DU68 A0A0L7L6X0 T1ICS4 A0A1B6I437 N6T8U6 A0A1B6G2Y9 U4UJI2 A0A0K8SGZ5 K7J7J5 A0A0J7KZ01 A0A0L7QS87 A0A1W4XHL4 A0A154PP36 A0A0C9REM8 A0A0C9RAV1 E2AEK8 E2BYV9 A0A151WJV4 A0A195EX02 A0A151IWW1 A0A195BTK6 A0A158NNW9 A0A084VMQ9 A0A151I956 F4WV82 A0A3L8DKY6 Q17BX4 E9J061 A0A1L8DZ29 A0A1Q3FHM4 A0A1Q3FI21 A0A1I8PSL0 A0A0P4W1G9 A0A182NB72 A0A1Q3FGK6 A0A023EVD3 A0A182JUD3 A0A1Y1KZ03 A0A182RA72 A0A182J7F3 A0A1B0DB09 A0A182X4Z8 A0A1B0GKL3 A0A182Y8I9 A0A182GZ27 A0A0L0CK58 A0A2M4A3R3 A0A1A9ZX86 W5J382 A0A182QF10 A0A182KWM7 A0A1B0C593 A0A182SNB6 A0A182VFM0 A0A182PUH8 A0A2M4A486 D3TR49 A0A1S4HD25 A0A182TFN3 A0A1I8JU27 V4BBD4 B0XAD1 A0A182GT47 A0A182MJ79 U5EKH4 A0A1A9Y1X5 A0A2M4A4K7 A0A336M2Q0 A0A226EQX8 A0A2M3Z1C8 A0A182VTL8 A0A1Y9G9M4 A0A2G8KXD8 A0A1A9UIN8 A0A026WQW2 A0A1W4V026 A0A2M4BH60 A0A1D2MYK9
PDB
2UXW
E-value=3.13502e-167,
Score=1512
Ontologies
PATHWAY
GO
PANTHER
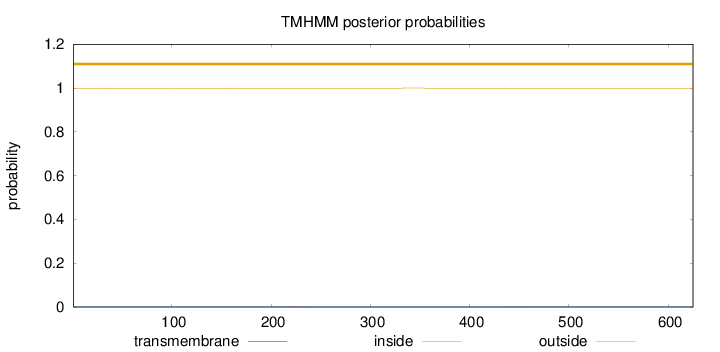
Topology
Length:
625
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0487499999999999
Exp number, first 60 AAs:
0.0044
Total prob of N-in:
0.00167
outside
1 - 625
Population Genetic Test Statistics
Pi
253.231341
Theta
198.428727
Tajima's D
0.797756
CLR
0.500714
CSRT
0.608219589020549
Interpretation
Uncertain
