Gene
KWMTBOMO02351
Pre Gene Modal
BGIBMGA014411
Annotation
PREDICTED:_DNA_repair_protein_complementing_XP-A_cells_homolog_[Papilio_machaon]
Full name
DNA repair protein complementing XP-A cells homolog
Alternative Name
Xeroderma pigmentosum group A-complementing protein homolog
Location in the cell
Nuclear Reliability : 2.484
Sequence
CDS
ATGGACAGCGATGAAGTAAACATATCCCAAGAGAAAAAAAACAAATTAACTGCCACACAGCTTGCCCGCATCGAACGGCAACGTCTACGGGCCAGAGCACTGCGTGACTCTCGACCCTTGCTGAGAAATGAAGATAAACCCGCACCAGTACCCACTCTTGGATGCGGAGGTGGATTCCTGCCAGAAGAGACACACGAGCTGAATGCAGTGGTTGCGCCTTGCGCTCCCCTCGTGCACCCCGGCCACCAACCCCATTGTCTTCATTGCGGACGACCCTTTCCACAATCTTTCCTCTATGACAACTTTGACTATTCAATTTGTGACGAGTGTAGGGATACTGAAGGAGAACACGCTCTCATGACACGCACAGAGGCCAAGAACGAGTACCTTTTGAAGGATTGCGACTTGGACAGTCGAGAACCCCCTTTGCGATGTTTGCGACGCCGCAACCCGCACAGCGCTCGGTTCGGCGACATGCGGTTGTACCTGCGCGCGCAGCTGCAGGAGCGCGCGCTGCAGGTGTGGGGCTCGCACGCGGCGCTGGACGAGGAGCGGGCGCGGCGGGAGGGCGCGCGGGCGGCGGGGGCGGGGCGGCGCGCAGTCCGCCGCCTGCGCGCGCTGCGGATGGACGTGCGCTCGAGTCTGTACACGCCCCAGCGCGCCGCACACGAGCACGAGTTCGGTCCCGAGACGCACGATGCCCGCGCCGACGTCTACTCCCGCTCCTGCCTCTCCTGCGGACACACCCAGTCCTATGAGAAGATGTGA
Protein
MDSDEVNISQEKKNKLTATQLARIERQRLRARALRDSRPLLRNEDKPAPVPTLGCGGGFLPEETHELNAVVAPCAPLVHPGHQPHCLHCGRPFPQSFLYDNFDYSICDECRDTEGEHALMTRTEAKNEYLLKDCDLDSREPPLRCLRRRNPHSARFGDMRLYLRAQLQERALQVWGSHAALDEERARREGARAAGAGRRAVRRLRALRMDVRSSLYTPQRAAHEHEFGPETHDARADVYSRSCLSCGHTQSYEKM
Summary
Description
Involved in DNA excision repair. Initiates repair by binding to damaged sites with various affinities, depending on the photoproduct and the transcriptional state of the region (By similarity).
Similarity
Belongs to the XPA family.
Keywords
Complete proteome
DNA damage
DNA repair
DNA-binding
Metal-binding
Nucleus
Reference proteome
Zinc
Zinc-finger
Feature
chain DNA repair protein complementing XP-A cells homolog
Uniprot
A0A2H1W8R9
A0A0N1IF35
A0A212FAP1
A0A1I8NXF5
A0A1B6FML9
B4JNH0
+ More
A0A1I8M3I0 A0A1W4UPD1 Q29I43 A0A0L0C301 B4HA92 B4M7L9 A0A182KCR8 A0A3P8W0I1 A0A1S3IZ50 A0A1S3IZN8 A0A182MIZ7 A0A182WIN9 A0A0Q9WEY0 A0A1B6JD02 A0A2Z5TRL6 A0A1B6L8J1 A0A182XZ33 A0A2P2I554 A0A1A9WHV3 W8C6W9 B4Q0F6 B3MSL8 A0A3B0JDJ3 A0A212EW90 H0RNH2 P28518 W5JI56 D3PFI1 A0A1A8EU74 B4I126 A0A0K8TVY6 A0A182IZW4 A0A0L7RBJ7 A0A1A8CYN2 A0A1A8B7Z2 A0A1A8JHS4 A0A034UX17 A0A2I4CG41 B3NSU2 W5JUE2 A0A2A3EUZ7 A0A1A8EBS3 A0A1A8RBW5 A0A154PMY7 E0VXW3 A0A1A8RGG6 A0A182P3R2 A0A182UQY2 A0A2M4BWM6 A0A0A1X2R5 A0A2J7R3S2 A0A1A8MEF5 A0A2M4BWH4 A0A182TYM8 A0A182I3I5 A0A182X3I5 A0A310SI10 A0A0M5J178 A0A0A1X1X9 A0A1A9ZRE5 B4L6M6 A0A088AJ88 A0A182KRM2 C3YQX9 I3KH57 A0A1B0FY38 D6WKJ8 A0A2T7NEJ7 A0A084VEZ3 A0A067QWU0 A0A1S4EEH0 A0A3B4EVF8 Q7QJL3 A0A2G8LLN3 A0A3P8N6D4 A0A1A9V625 A0A182NH84 A0A1A7XNW5 A0A3P9I8T6 A0A182R4Q7 A0A1A7Y0B6 A0A3P9B0M4 U5EXT0 A0A147A513 A0A0S7LP57 E2B616 A0A3B3ZEZ4 A0A182QNJ1 A0A151HYY1 A0A1W4X6C9 A0A158NWB2 H3BX47 H3DPW4 F4X4S6
A0A1I8M3I0 A0A1W4UPD1 Q29I43 A0A0L0C301 B4HA92 B4M7L9 A0A182KCR8 A0A3P8W0I1 A0A1S3IZ50 A0A1S3IZN8 A0A182MIZ7 A0A182WIN9 A0A0Q9WEY0 A0A1B6JD02 A0A2Z5TRL6 A0A1B6L8J1 A0A182XZ33 A0A2P2I554 A0A1A9WHV3 W8C6W9 B4Q0F6 B3MSL8 A0A3B0JDJ3 A0A212EW90 H0RNH2 P28518 W5JI56 D3PFI1 A0A1A8EU74 B4I126 A0A0K8TVY6 A0A182IZW4 A0A0L7RBJ7 A0A1A8CYN2 A0A1A8B7Z2 A0A1A8JHS4 A0A034UX17 A0A2I4CG41 B3NSU2 W5JUE2 A0A2A3EUZ7 A0A1A8EBS3 A0A1A8RBW5 A0A154PMY7 E0VXW3 A0A1A8RGG6 A0A182P3R2 A0A182UQY2 A0A2M4BWM6 A0A0A1X2R5 A0A2J7R3S2 A0A1A8MEF5 A0A2M4BWH4 A0A182TYM8 A0A182I3I5 A0A182X3I5 A0A310SI10 A0A0M5J178 A0A0A1X1X9 A0A1A9ZRE5 B4L6M6 A0A088AJ88 A0A182KRM2 C3YQX9 I3KH57 A0A1B0FY38 D6WKJ8 A0A2T7NEJ7 A0A084VEZ3 A0A067QWU0 A0A1S4EEH0 A0A3B4EVF8 Q7QJL3 A0A2G8LLN3 A0A3P8N6D4 A0A1A9V625 A0A182NH84 A0A1A7XNW5 A0A3P9I8T6 A0A182R4Q7 A0A1A7Y0B6 A0A3P9B0M4 U5EXT0 A0A147A513 A0A0S7LP57 E2B616 A0A3B3ZEZ4 A0A182QNJ1 A0A151HYY1 A0A1W4X6C9 A0A158NWB2 H3BX47 H3DPW4 F4X4S6
Pubmed
26354079
22118469
17994087
25315136
15632085
26108605
+ More
24487278 26760975 25244985 24495485 17550304 1764072 7673233 10731132 12537572 10731137 20920257 23761445 25348373 20566863 25830018 20966253 18563158 25186727 18362917 19820115 24438588 24845553 12364791 14747013 17210077 29023486 17554307 20798317 25463417 21347285 15496914 21719571
24487278 26760975 25244985 24495485 17550304 1764072 7673233 10731132 12537572 10731137 20920257 23761445 25348373 20566863 25830018 20966253 18563158 25186727 18362917 19820115 24438588 24845553 12364791 14747013 17210077 29023486 17554307 20798317 25463417 21347285 15496914 21719571
EMBL
ODYU01007043
SOQ49460.1
KQ460401
KPJ14970.1
AGBW02009439
OWR50806.1
+ More
GECZ01018347 JAS51422.1 CH916371 EDV92263.1 CH379064 EAL31563.1 JRES01000960 KNC26728.1 CH479239 EDW37486.1 CH940653 EDW62786.1 AXCM01007945 KRF80791.1 GECU01019471 GECU01010708 JAS88235.1 JAS96998.1 FX985852 BBA93739.1 GEBQ01020022 JAT19955.1 IACF01003544 LAB69158.1 GAMC01000887 JAC05669.1 CM000162 EDX01240.1 CH902622 EDV34773.1 OUUW01000003 SPP78202.1 AGBW02012041 OWR45762.1 BT132894 AEV23918.1 D31893 D31892 AE014298 AL033125 ADMH02001474 ETN62454.1 BT120387 ADD16467.1 HAEB01003880 SBQ50407.1 CH480819 EDW53207.1 GDHF01033675 JAI18639.1 KQ414617 KOC68239.1 HADZ01020009 SBP83950.1 HADY01024130 HAEJ01016854 SBP62615.1 HAED01022720 HAEE01005649 SBR09487.1 GAKP01023437 JAC35521.1 CH954180 EDV45772.1 ADMH02000120 ETN67766.1 KZ288185 PBC34969.1 HAEA01014571 SBQ43051.1 HAEH01015603 SBS03172.1 KQ434977 KZC12824.1 DS235841 EEB18219.1 HAEI01008350 SBS04777.1 GGFJ01008335 MBW57476.1 GBXI01008663 JAD05629.1 NEVH01007818 PNF35483.1 HAEF01014023 HAEG01007645 SBR55182.1 GGFJ01008268 MBW57409.1 APCN01000213 KQ768636 OAD53092.1 CP012528 ALC49268.1 GBXI01009614 JAD04678.1 CH933812 EDW06022.1 GG666544 EEN57320.1 AERX01023435 AJVK01031199 KQ971342 EFA03569.1 PZQS01000013 PVD19565.1 ATLV01012357 KE524785 KFB36537.1 KK852921 KDR13782.1 AAAB01008807 EAA04751.3 MRZV01000040 PIK61145.1 HADW01018288 SBP19688.1 HADX01001534 SBP23766.1 GANO01002389 JAB57482.1 GCES01012717 JAR73606.1 GBYX01157920 GBYX01157915 GBYX01157914 GBYX01157913 JAO91272.1 GL445900 EFN88877.1 AXCN02000893 KQ976730 KYM76453.1 ADTU01027864 GL888679 EGI58499.1
GECZ01018347 JAS51422.1 CH916371 EDV92263.1 CH379064 EAL31563.1 JRES01000960 KNC26728.1 CH479239 EDW37486.1 CH940653 EDW62786.1 AXCM01007945 KRF80791.1 GECU01019471 GECU01010708 JAS88235.1 JAS96998.1 FX985852 BBA93739.1 GEBQ01020022 JAT19955.1 IACF01003544 LAB69158.1 GAMC01000887 JAC05669.1 CM000162 EDX01240.1 CH902622 EDV34773.1 OUUW01000003 SPP78202.1 AGBW02012041 OWR45762.1 BT132894 AEV23918.1 D31893 D31892 AE014298 AL033125 ADMH02001474 ETN62454.1 BT120387 ADD16467.1 HAEB01003880 SBQ50407.1 CH480819 EDW53207.1 GDHF01033675 JAI18639.1 KQ414617 KOC68239.1 HADZ01020009 SBP83950.1 HADY01024130 HAEJ01016854 SBP62615.1 HAED01022720 HAEE01005649 SBR09487.1 GAKP01023437 JAC35521.1 CH954180 EDV45772.1 ADMH02000120 ETN67766.1 KZ288185 PBC34969.1 HAEA01014571 SBQ43051.1 HAEH01015603 SBS03172.1 KQ434977 KZC12824.1 DS235841 EEB18219.1 HAEI01008350 SBS04777.1 GGFJ01008335 MBW57476.1 GBXI01008663 JAD05629.1 NEVH01007818 PNF35483.1 HAEF01014023 HAEG01007645 SBR55182.1 GGFJ01008268 MBW57409.1 APCN01000213 KQ768636 OAD53092.1 CP012528 ALC49268.1 GBXI01009614 JAD04678.1 CH933812 EDW06022.1 GG666544 EEN57320.1 AERX01023435 AJVK01031199 KQ971342 EFA03569.1 PZQS01000013 PVD19565.1 ATLV01012357 KE524785 KFB36537.1 KK852921 KDR13782.1 AAAB01008807 EAA04751.3 MRZV01000040 PIK61145.1 HADW01018288 SBP19688.1 HADX01001534 SBP23766.1 GANO01002389 JAB57482.1 GCES01012717 JAR73606.1 GBYX01157920 GBYX01157915 GBYX01157914 GBYX01157913 JAO91272.1 GL445900 EFN88877.1 AXCN02000893 KQ976730 KYM76453.1 ADTU01027864 GL888679 EGI58499.1
Proteomes
UP000053240
UP000007151
UP000095300
UP000001070
UP000095301
UP000192221
+ More
UP000001819 UP000037069 UP000008744 UP000008792 UP000075881 UP000265120 UP000085678 UP000075883 UP000075920 UP000076408 UP000091820 UP000002282 UP000007801 UP000268350 UP000000803 UP000000673 UP000001292 UP000075880 UP000053825 UP000192220 UP000008711 UP000242457 UP000076502 UP000009046 UP000075885 UP000075903 UP000235965 UP000075902 UP000075840 UP000076407 UP000092553 UP000092445 UP000009192 UP000005203 UP000075882 UP000001554 UP000005207 UP000092462 UP000007266 UP000245119 UP000030765 UP000027135 UP000079169 UP000261460 UP000007062 UP000230750 UP000265100 UP000078200 UP000075884 UP000265200 UP000075900 UP000265160 UP000265000 UP000008237 UP000261520 UP000075886 UP000078540 UP000192223 UP000005205 UP000007303 UP000007755
UP000001819 UP000037069 UP000008744 UP000008792 UP000075881 UP000265120 UP000085678 UP000075883 UP000075920 UP000076408 UP000091820 UP000002282 UP000007801 UP000268350 UP000000803 UP000000673 UP000001292 UP000075880 UP000053825 UP000192220 UP000008711 UP000242457 UP000076502 UP000009046 UP000075885 UP000075903 UP000235965 UP000075902 UP000075840 UP000076407 UP000092553 UP000092445 UP000009192 UP000005203 UP000075882 UP000001554 UP000005207 UP000092462 UP000007266 UP000245119 UP000030765 UP000027135 UP000079169 UP000261460 UP000007062 UP000230750 UP000265100 UP000078200 UP000075884 UP000265200 UP000075900 UP000265160 UP000265000 UP000008237 UP000261520 UP000075886 UP000078540 UP000192223 UP000005205 UP000007303 UP000007755
PRIDE
Interpro
SUPFAM
SSF46955
SSF46955
Gene 3D
ProteinModelPortal
A0A2H1W8R9
A0A0N1IF35
A0A212FAP1
A0A1I8NXF5
A0A1B6FML9
B4JNH0
+ More
A0A1I8M3I0 A0A1W4UPD1 Q29I43 A0A0L0C301 B4HA92 B4M7L9 A0A182KCR8 A0A3P8W0I1 A0A1S3IZ50 A0A1S3IZN8 A0A182MIZ7 A0A182WIN9 A0A0Q9WEY0 A0A1B6JD02 A0A2Z5TRL6 A0A1B6L8J1 A0A182XZ33 A0A2P2I554 A0A1A9WHV3 W8C6W9 B4Q0F6 B3MSL8 A0A3B0JDJ3 A0A212EW90 H0RNH2 P28518 W5JI56 D3PFI1 A0A1A8EU74 B4I126 A0A0K8TVY6 A0A182IZW4 A0A0L7RBJ7 A0A1A8CYN2 A0A1A8B7Z2 A0A1A8JHS4 A0A034UX17 A0A2I4CG41 B3NSU2 W5JUE2 A0A2A3EUZ7 A0A1A8EBS3 A0A1A8RBW5 A0A154PMY7 E0VXW3 A0A1A8RGG6 A0A182P3R2 A0A182UQY2 A0A2M4BWM6 A0A0A1X2R5 A0A2J7R3S2 A0A1A8MEF5 A0A2M4BWH4 A0A182TYM8 A0A182I3I5 A0A182X3I5 A0A310SI10 A0A0M5J178 A0A0A1X1X9 A0A1A9ZRE5 B4L6M6 A0A088AJ88 A0A182KRM2 C3YQX9 I3KH57 A0A1B0FY38 D6WKJ8 A0A2T7NEJ7 A0A084VEZ3 A0A067QWU0 A0A1S4EEH0 A0A3B4EVF8 Q7QJL3 A0A2G8LLN3 A0A3P8N6D4 A0A1A9V625 A0A182NH84 A0A1A7XNW5 A0A3P9I8T6 A0A182R4Q7 A0A1A7Y0B6 A0A3P9B0M4 U5EXT0 A0A147A513 A0A0S7LP57 E2B616 A0A3B3ZEZ4 A0A182QNJ1 A0A151HYY1 A0A1W4X6C9 A0A158NWB2 H3BX47 H3DPW4 F4X4S6
A0A1I8M3I0 A0A1W4UPD1 Q29I43 A0A0L0C301 B4HA92 B4M7L9 A0A182KCR8 A0A3P8W0I1 A0A1S3IZ50 A0A1S3IZN8 A0A182MIZ7 A0A182WIN9 A0A0Q9WEY0 A0A1B6JD02 A0A2Z5TRL6 A0A1B6L8J1 A0A182XZ33 A0A2P2I554 A0A1A9WHV3 W8C6W9 B4Q0F6 B3MSL8 A0A3B0JDJ3 A0A212EW90 H0RNH2 P28518 W5JI56 D3PFI1 A0A1A8EU74 B4I126 A0A0K8TVY6 A0A182IZW4 A0A0L7RBJ7 A0A1A8CYN2 A0A1A8B7Z2 A0A1A8JHS4 A0A034UX17 A0A2I4CG41 B3NSU2 W5JUE2 A0A2A3EUZ7 A0A1A8EBS3 A0A1A8RBW5 A0A154PMY7 E0VXW3 A0A1A8RGG6 A0A182P3R2 A0A182UQY2 A0A2M4BWM6 A0A0A1X2R5 A0A2J7R3S2 A0A1A8MEF5 A0A2M4BWH4 A0A182TYM8 A0A182I3I5 A0A182X3I5 A0A310SI10 A0A0M5J178 A0A0A1X1X9 A0A1A9ZRE5 B4L6M6 A0A088AJ88 A0A182KRM2 C3YQX9 I3KH57 A0A1B0FY38 D6WKJ8 A0A2T7NEJ7 A0A084VEZ3 A0A067QWU0 A0A1S4EEH0 A0A3B4EVF8 Q7QJL3 A0A2G8LLN3 A0A3P8N6D4 A0A1A9V625 A0A182NH84 A0A1A7XNW5 A0A3P9I8T6 A0A182R4Q7 A0A1A7Y0B6 A0A3P9B0M4 U5EXT0 A0A147A513 A0A0S7LP57 E2B616 A0A3B3ZEZ4 A0A182QNJ1 A0A151HYY1 A0A1W4X6C9 A0A158NWB2 H3BX47 H3DPW4 F4X4S6
PDB
1D4U
E-value=3.67206e-27,
Score=300
Ontologies
GO
PANTHER
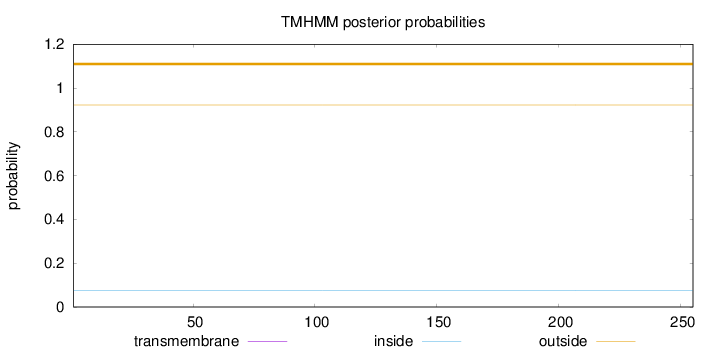
Topology
Subcellular location
Nucleus
Length:
255
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07689
outside
1 - 255
Population Genetic Test Statistics
Pi
17.676903
Theta
5.715372
Tajima's D
-2.274518
CLR
348.074641
CSRT
0.00249987500624969
Interpretation
Possibly Positive selection
