Gene
KWMTBOMO02350
Pre Gene Modal
BGIBMGA014410
Annotation
PREDICTED:_uncharacterized_protein_LOC106718269_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.327 Extracellular Reliability : 1.266
Sequence
CDS
ATGAAAGCAAACGGCTGGAGCAAGTCACATGTTATGACGGCGCTTACTTTGGGACTCCGTGAACAAGCTTTGACCATCCTGGAAACCTTAGGAAAGGATGCAACCTATGAGCAGCTCGTGGAAGCGTTGGAGTCCAGATATAGAGACGCGCATCTGGAACACGTGTTTCGTGCCCAGTTGAAGGAACGAGTGCAGCGCGGTAACGAGAACCTCCCACAGTGGGCTTTCGAGGTTGAGAAGTTAGTCGGCAAGGCGAACCGCTCTTCACCTGCCCTTATTGATGGTAACCTGGTGCAAGCATTCATCGACGGTATCCGAGACTCCAGTCAGAGCTGCGGTGCGATTGGGTCATCACGAGGCCCTTAA
Protein
MKANGWSKSHVMTALTLGLREQALTILETLGKDATYEQLVEALESRYRDAHLEHVFRAQLKERVQRGNENLPQWAFEVEKLVGKANRSSPALIDGNLVQAFIDGIRDSSQSCGAIGSSRGP
Summary
Uniprot
H9JXZ2
A0A2S4PKQ1
A0A1Y1KUJ8
A0A2C9KZC3
T1EMU9
T1F1S0
+ More
T1FBT1 T1FLB2 A0A034V2G1 T1ERN8 A0A1B6EBV0 T1FLI2 T1FDM1 A0A1B6EED3 T1F1N1 T1EUD3 A0A3Q0ITM1 T1FIB4 T1FFS5 A0A310SNN0 T1FFF5 T1FGP7 T1F8U0 A0A023EWI1 T1FJK5 A0A0A9W5I2 T1FFW2 T1EUN5 T1F1D1 T1EPR1 T1FLI8 T1FBS8 A0A1B6HG85 T1F6D3 A0A1B6K8J8 T1FEQ7 A0A1B6MHK7 T1FI89 T1F7U7 T1EV56 T1EQZ4 T1FFV1 T1ERI3 T1F7X8 T1PPT0 A0A2H9T5P4 A0A2J7REY1 T1F9L6 T1EUG7 T1FA86 K9LCE4 A0A087U818 A0A2H9T5I1 A0A034VYS8 A0A1B6LVC0 A0A087TBQ9 A0A0J7JVT9 A0A1B6KFC4 A0A1B6KPQ6 A0A087UIG5 A0A087TBY4 A0A0A9YDH8 A0A0L0BW24 A0A087SUZ9 A0A1B6KFC1 A0A1B6KAB4 A0A087T3I4 A0A2J7PND1 A0A1B6LDH1 W8AVY4 W8AJP1 A0A2J7R0R8 A0A2G8KDS6 A0A1A9VE53 A0A0A9Y0N5 A0A2G8JMV7 A0A0A1WD72 A0A2G8JS73 A0A0K8SSG3 A0A0K8TH95 V4AEN8 A0A260YMM9 A0A087UEM8 K7JZZ1 A0A0A9YIJ1 A0A1A9ZSE9
T1FBT1 T1FLB2 A0A034V2G1 T1ERN8 A0A1B6EBV0 T1FLI2 T1FDM1 A0A1B6EED3 T1F1N1 T1EUD3 A0A3Q0ITM1 T1FIB4 T1FFS5 A0A310SNN0 T1FFF5 T1FGP7 T1F8U0 A0A023EWI1 T1FJK5 A0A0A9W5I2 T1FFW2 T1EUN5 T1F1D1 T1EPR1 T1FLI8 T1FBS8 A0A1B6HG85 T1F6D3 A0A1B6K8J8 T1FEQ7 A0A1B6MHK7 T1FI89 T1F7U7 T1EV56 T1EQZ4 T1FFV1 T1ERI3 T1F7X8 T1PPT0 A0A2H9T5P4 A0A2J7REY1 T1F9L6 T1EUG7 T1FA86 K9LCE4 A0A087U818 A0A2H9T5I1 A0A034VYS8 A0A1B6LVC0 A0A087TBQ9 A0A0J7JVT9 A0A1B6KFC4 A0A1B6KPQ6 A0A087UIG5 A0A087TBY4 A0A0A9YDH8 A0A0L0BW24 A0A087SUZ9 A0A1B6KFC1 A0A1B6KAB4 A0A087T3I4 A0A2J7PND1 A0A1B6LDH1 W8AVY4 W8AJP1 A0A2J7R0R8 A0A2G8KDS6 A0A1A9VE53 A0A0A9Y0N5 A0A2G8JMV7 A0A0A1WD72 A0A2G8JS73 A0A0K8SSG3 A0A0K8TH95 V4AEN8 A0A260YMM9 A0A087UEM8 K7JZZ1 A0A0A9YIJ1 A0A1A9ZSE9
Pubmed
EMBL
BABH01041732
PEDP01002624
POS82547.1
GEZM01079005
JAV62537.1
AMQM01000050
+ More
KB095811 ESO12076.1 AMQM01003259 KB096080 ESO08469.1 AMQM01006081 KB097269 ESN97863.1 AMQM01010234 KB097577 ESN94025.1 GAKP01022625 JAC36327.1 AMQM01000852 KB096742 ESO02334.1 GEDC01001886 JAS35412.1 AMQM01010829 KB095936 ESO09675.1 AMQM01006535 KB097487 ESN96915.1 GEDC01001027 JAS36271.1 AMQM01003246 ESO08416.1 AMQM01001433 KB097495 ESN96614.1 AMQM01008234 AMQM01008235 KB097744 ESN90815.1 AMQM01007172 KB097579 ESN93936.1 KQ762552 OAD55663.1 AMQM01007080 KB097563 ESN94735.1 AMQM01007491 KB097628 ESN93232.1 AMQM01005140 KB096830 ESO01055.1 GBBI01005037 JAC13675.1 AMQM01008746 AMQM01008747 KB095840 ESO11223.1 GBHO01043510 GBHO01043509 GBHO01043507 GBHO01043506 JAG00094.1 JAG00095.1 JAG00097.1 JAG00098.1 AMQM01007184 ESN93988.1 AMQM01001501 KB097571 ESN94169.1 AMQM01003171 KB096023 ESO09130.1 AMQM01000492 AMQM01000493 KB096324 ESO05975.1 AMQM01010853 KB095953 ESO09617.1 AMQM01006079 ESN97860.1 GECU01034034 JAS73672.1 AMQM01004449 KB096551 ESO04038.1 GECU01000203 JAT07504.1 AMQM01006849 KB097528 ESN95383.1 GEBQ01004606 JAT35371.1 AMQM01008217 KB097739 ESN90891.1 AMQM01004859 KB096716 ESO02813.1 AMQM01001610 AMQM01001611 ESN94407.1 AMQM01000727 AMQM01000728 ESO01826.1 AMQM01007178 ESN93966.1 AMQM01000828 ESO02231.1 AMQM01004874 ESO02851.1 KA650088 AFP64717.1 NSIT01000165 PJE78522.1 NEVH01004423 PNF39393.1 AMQM01005413 KB096945 ESO00339.1 AMQM01001452 ESN96681.1 AMQM01005564 KB096983 ESO00299.1 JF835027 AFH54163.1 KK118640 KFM73507.1 NSIT01000173 PJE78480.1 GAKP01011740 JAC47212.1 GEBQ01012350 JAT27627.1 KK114486 KFM62548.1 LBMM01026972 KMQ82254.1 GEBQ01029848 JAT10129.1 GEBQ01026576 JAT13401.1 KK119952 KFM77154.1 KK114513 KFM62623.1 GBHO01014451 JAG29153.1 JRES01001250 KNC24232.1 KK112080 KFM56688.1 GEBQ01029815 JAT10162.1 GEBQ01031607 JAT08370.1 KK113242 KFM59673.1 NEVH01023954 PNF17836.1 GEBQ01018473 JAT21504.1 GAMC01021624 JAB84931.1 GAMC01021622 JAB84933.1 NEVH01008218 PNF34431.1 MRZV01000662 PIK46151.1 GBHO01020524 GBRD01015929 JAG23080.1 JAG49897.1 MRZV01001557 PIK37106.1 GBXI01017626 JAC96665.1 MRZV01001335 PIK38624.1 GBRD01009708 JAG56116.1 GBRD01000849 JAG64972.1 KB202199 ESO91816.1 NIPN01001566 OZF74667.1 KK119487 KFM75817.1 AAZX01013784 GBHO01014259 JAG29345.1
KB095811 ESO12076.1 AMQM01003259 KB096080 ESO08469.1 AMQM01006081 KB097269 ESN97863.1 AMQM01010234 KB097577 ESN94025.1 GAKP01022625 JAC36327.1 AMQM01000852 KB096742 ESO02334.1 GEDC01001886 JAS35412.1 AMQM01010829 KB095936 ESO09675.1 AMQM01006535 KB097487 ESN96915.1 GEDC01001027 JAS36271.1 AMQM01003246 ESO08416.1 AMQM01001433 KB097495 ESN96614.1 AMQM01008234 AMQM01008235 KB097744 ESN90815.1 AMQM01007172 KB097579 ESN93936.1 KQ762552 OAD55663.1 AMQM01007080 KB097563 ESN94735.1 AMQM01007491 KB097628 ESN93232.1 AMQM01005140 KB096830 ESO01055.1 GBBI01005037 JAC13675.1 AMQM01008746 AMQM01008747 KB095840 ESO11223.1 GBHO01043510 GBHO01043509 GBHO01043507 GBHO01043506 JAG00094.1 JAG00095.1 JAG00097.1 JAG00098.1 AMQM01007184 ESN93988.1 AMQM01001501 KB097571 ESN94169.1 AMQM01003171 KB096023 ESO09130.1 AMQM01000492 AMQM01000493 KB096324 ESO05975.1 AMQM01010853 KB095953 ESO09617.1 AMQM01006079 ESN97860.1 GECU01034034 JAS73672.1 AMQM01004449 KB096551 ESO04038.1 GECU01000203 JAT07504.1 AMQM01006849 KB097528 ESN95383.1 GEBQ01004606 JAT35371.1 AMQM01008217 KB097739 ESN90891.1 AMQM01004859 KB096716 ESO02813.1 AMQM01001610 AMQM01001611 ESN94407.1 AMQM01000727 AMQM01000728 ESO01826.1 AMQM01007178 ESN93966.1 AMQM01000828 ESO02231.1 AMQM01004874 ESO02851.1 KA650088 AFP64717.1 NSIT01000165 PJE78522.1 NEVH01004423 PNF39393.1 AMQM01005413 KB096945 ESO00339.1 AMQM01001452 ESN96681.1 AMQM01005564 KB096983 ESO00299.1 JF835027 AFH54163.1 KK118640 KFM73507.1 NSIT01000173 PJE78480.1 GAKP01011740 JAC47212.1 GEBQ01012350 JAT27627.1 KK114486 KFM62548.1 LBMM01026972 KMQ82254.1 GEBQ01029848 JAT10129.1 GEBQ01026576 JAT13401.1 KK119952 KFM77154.1 KK114513 KFM62623.1 GBHO01014451 JAG29153.1 JRES01001250 KNC24232.1 KK112080 KFM56688.1 GEBQ01029815 JAT10162.1 GEBQ01031607 JAT08370.1 KK113242 KFM59673.1 NEVH01023954 PNF17836.1 GEBQ01018473 JAT21504.1 GAMC01021624 JAB84931.1 GAMC01021622 JAB84933.1 NEVH01008218 PNF34431.1 MRZV01000662 PIK46151.1 GBHO01020524 GBRD01015929 JAG23080.1 JAG49897.1 MRZV01001557 PIK37106.1 GBXI01017626 JAC96665.1 MRZV01001335 PIK38624.1 GBRD01009708 JAG56116.1 GBRD01000849 JAG64972.1 KB202199 ESO91816.1 NIPN01001566 OZF74667.1 KK119487 KFM75817.1 AAZX01013784 GBHO01014259 JAG29345.1
Proteomes
PRIDE
Pfam
Interpro
IPR036397
RNaseH_sf
+ More
IPR041588 Integrase_H2C2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR000571 Znf_CCCH
IPR036875 Znf_CCHC_sf
IPR002589 Macro_dom
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR041577 RT_RNaseH_2
IPR000477 RT_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR005162 Retrotrans_gag_dom
IPR036361 SAP_dom_sf
IPR000727 T_SNARE_dom
IPR003034 SAP_dom
IPR036119 NOS_N_sf
IPR029039 Flavoprotein-like_sf
IPR008254 Flavodoxin/NO_synth
IPR004030 NOS_N
IPR002110 Ankyrin_rpt
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR008906 HATC_C_dom
IPR005312 DUF1759
IPR026523 PNMA
IPR002492 Transposase_Tc1-like
IPR009057 Homeobox-like_sf
IPR038717 Tc1-like_DDE_dom
IPR041588 Integrase_H2C2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR000571 Znf_CCCH
IPR036875 Znf_CCHC_sf
IPR002589 Macro_dom
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR041577 RT_RNaseH_2
IPR000477 RT_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR005162 Retrotrans_gag_dom
IPR036361 SAP_dom_sf
IPR000727 T_SNARE_dom
IPR003034 SAP_dom
IPR036119 NOS_N_sf
IPR029039 Flavoprotein-like_sf
IPR008254 Flavodoxin/NO_synth
IPR004030 NOS_N
IPR002110 Ankyrin_rpt
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR008906 HATC_C_dom
IPR005312 DUF1759
IPR026523 PNMA
IPR002492 Transposase_Tc1-like
IPR009057 Homeobox-like_sf
IPR038717 Tc1-like_DDE_dom
SUPFAM
CDD
ProteinModelPortal
H9JXZ2
A0A2S4PKQ1
A0A1Y1KUJ8
A0A2C9KZC3
T1EMU9
T1F1S0
+ More
T1FBT1 T1FLB2 A0A034V2G1 T1ERN8 A0A1B6EBV0 T1FLI2 T1FDM1 A0A1B6EED3 T1F1N1 T1EUD3 A0A3Q0ITM1 T1FIB4 T1FFS5 A0A310SNN0 T1FFF5 T1FGP7 T1F8U0 A0A023EWI1 T1FJK5 A0A0A9W5I2 T1FFW2 T1EUN5 T1F1D1 T1EPR1 T1FLI8 T1FBS8 A0A1B6HG85 T1F6D3 A0A1B6K8J8 T1FEQ7 A0A1B6MHK7 T1FI89 T1F7U7 T1EV56 T1EQZ4 T1FFV1 T1ERI3 T1F7X8 T1PPT0 A0A2H9T5P4 A0A2J7REY1 T1F9L6 T1EUG7 T1FA86 K9LCE4 A0A087U818 A0A2H9T5I1 A0A034VYS8 A0A1B6LVC0 A0A087TBQ9 A0A0J7JVT9 A0A1B6KFC4 A0A1B6KPQ6 A0A087UIG5 A0A087TBY4 A0A0A9YDH8 A0A0L0BW24 A0A087SUZ9 A0A1B6KFC1 A0A1B6KAB4 A0A087T3I4 A0A2J7PND1 A0A1B6LDH1 W8AVY4 W8AJP1 A0A2J7R0R8 A0A2G8KDS6 A0A1A9VE53 A0A0A9Y0N5 A0A2G8JMV7 A0A0A1WD72 A0A2G8JS73 A0A0K8SSG3 A0A0K8TH95 V4AEN8 A0A260YMM9 A0A087UEM8 K7JZZ1 A0A0A9YIJ1 A0A1A9ZSE9
T1FBT1 T1FLB2 A0A034V2G1 T1ERN8 A0A1B6EBV0 T1FLI2 T1FDM1 A0A1B6EED3 T1F1N1 T1EUD3 A0A3Q0ITM1 T1FIB4 T1FFS5 A0A310SNN0 T1FFF5 T1FGP7 T1F8U0 A0A023EWI1 T1FJK5 A0A0A9W5I2 T1FFW2 T1EUN5 T1F1D1 T1EPR1 T1FLI8 T1FBS8 A0A1B6HG85 T1F6D3 A0A1B6K8J8 T1FEQ7 A0A1B6MHK7 T1FI89 T1F7U7 T1EV56 T1EQZ4 T1FFV1 T1ERI3 T1F7X8 T1PPT0 A0A2H9T5P4 A0A2J7REY1 T1F9L6 T1EUG7 T1FA86 K9LCE4 A0A087U818 A0A2H9T5I1 A0A034VYS8 A0A1B6LVC0 A0A087TBQ9 A0A0J7JVT9 A0A1B6KFC4 A0A1B6KPQ6 A0A087UIG5 A0A087TBY4 A0A0A9YDH8 A0A0L0BW24 A0A087SUZ9 A0A1B6KFC1 A0A1B6KAB4 A0A087T3I4 A0A2J7PND1 A0A1B6LDH1 W8AVY4 W8AJP1 A0A2J7R0R8 A0A2G8KDS6 A0A1A9VE53 A0A0A9Y0N5 A0A2G8JMV7 A0A0A1WD72 A0A2G8JS73 A0A0K8SSG3 A0A0K8TH95 V4AEN8 A0A260YMM9 A0A087UEM8 K7JZZ1 A0A0A9YIJ1 A0A1A9ZSE9
Ontologies
GO
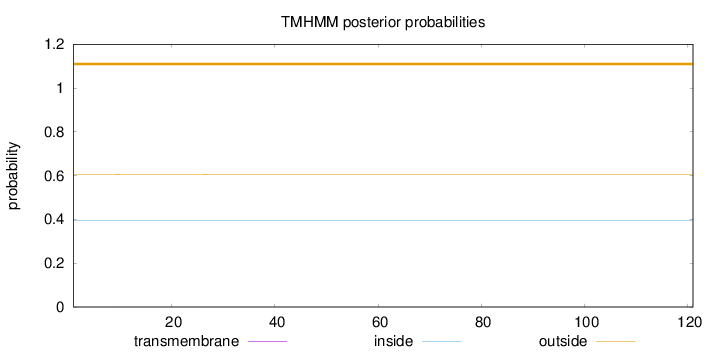
Topology
Length:
121
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.003
Exp number, first 60 AAs:
0.00286
Total prob of N-in:
0.39517
outside
1 - 121
Population Genetic Test Statistics
Pi
0.413969
Theta
4.04599
Tajima's D
-2.416374
CLR
405.922446
CSRT
0.00039998000099995
Interpretation
Possibly Positive selection
