Gene
KWMTBOMO02340
Pre Gene Modal
BGIBMGA003795
Annotation
PREDICTED:_nephrin-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 0.826
Sequence
CDS
ATGTTAGAAACCATAGAATGTTTGGTCTATATGGAAATCGAACCCACGACCCTCGACGCACCAAATTTTTCAAATAGCAAATGTTGTTACAGCTTCGACGTCCGCGGGAAGCCATTGTCAGAAGCCCGACATTGGTCCGCTCCGGAAGTGTTCGGTCCTCGAGCCCACTTCTCCACCTCCCCCGCCCCGGCGTCGCTGCTGGTCCGGGACGTGAAGCGCCGCGACGAGGGCGTCTACAGGTGCCGGGTGGATTTCCGCAACACGCAGACAACTAGTTTCCGCTACAATCTTACTGTTGTCGTGCCACCTGAGAAGCCAGTAGTGGTCGACAGGTGGGGTCGCGTAATCAACACTACGACCCTTGGTCCTCACGAGGAAGGTGATGACGTCCTCCTCACGTGCAGAGTACTCGGAGGTCGACCAGAGCCTTCGGTGCGTTGGCTGGTGAATGGAGTCCTTGTCGATGAGGAGTATGAGCACAATACTGGTGACGTCATAGAGAACCGACTACTATGGCCCGCGATACGACGCGCCGACTACGCAGCAGTCTTCACTTGCCAAGCTGCTAACTCGCATCTAGTGCCGCCCAAGGAGGTGTCGCTGGTACTGGACATGTTCTTGCGACCATTAACCGTAGAAATAAAGAAGCCTGCGGAAACGGGAGAAGGAGGAGTTCTAACAGCGGAACGGCGCTACGAAGTGGCCTGCGAGTCAGCGGGCAGCCGTCCCCCGGCCATCGTCACTTGGTTCAAGGGCAAGAGACAGCTGAAGAGGATAACGGAGGAGCTCCGGGACAACCTGACTATGAGCGTGATGAGCTTCGTTCCGACCCTGGATGATGACGGAAAACCAATCACATGTCGAGCCGAGAATCCTAACGTAACCACCCTATCTATGGAAACCTCATGGACTATCAACGTAGTCTACCCTCCAGTCGTCCGACTTCGCCTGGGCAGCTCGTTGGCAGCAGGAGACATCAAGGAGGGAGACGACGTCTACTTCGAGTGTCATGTGAGGGCCAACCCTCCTGCTAGGAAACTCTCCTGGCTACATGATGATCGTCCACTGGCTCACAACGCGAGTGCACGCGTGTTCCACAGCAACCAGAGCCTTGTGCTGCAGAAAGTGACGCGACACAGCAGCGGACGGTATGCCTGCTCTGCACTCAATGCAGAAGGAGAGACTGTATCTAATGAACTGCACTTTAGAGTTAAATATGCACCTTCCTGTCGAAGCGGCGGTGTGTCGGTGGTGGGTGCGGCCCGCGGTGAGTCCGTGGTAATAGTGTGCAAAGTGGACGCCGACCCCCCCGCCGCCGTCTTCAAGTGGAAGTTCAACAACTCCGGGGAGACCCTCGACGTCGCCGCCGACAGATACACCTCTAATGGCAGCGCCTCTAGCCTTAAGTACACTCCAGTAGCCGACCTGGACTATGGGACCCTGTCGTGCTCGGCGTCCAACGAGGTAGGCGCACAGCTCGCCCCCTGTGTGTTCCAGATGGTGGTGGCAGGCAAGCCGCACGCCCCGCGCAACTGCACGCTGTGGAATCAGACGGCGGAGTCGGTGGAGGTGTCGTGCGTGGCGGGCTTCGACGGCGGCCTGCCGCAGACCTTCCTGCTAGAGGTCTACCATGCCGACGGATCCTCCCCGAGAGTAAACTTAACATCAGAAGAGCCGTCGTGGACGATACGAGGATTGGAGTGGGACGTGCGCTTTCGACTCATAGCCGTGGCGGTGAACAGCAAGGGTCGATCACCACCCGCCTCCCTAGATGACGTGCTGTTCAGAGACCCGGAGAAGAGAACAGCAACGGAAGCGTCACTAGGCATGAGCAGCATGTTGGGCGCAGCGGGAGCGGGAGCGGGTGCGATATTCGCGCTGTTGGCGGGGGCGTGTGCCGTGTCGCTGCGACGGAGGCGGCGGCCGCGTCTAGCCAAGCCGCTGCCCGCGCCGCTCGTCACCCAGCACCTCACTCTTAAACAACCAGAATTGGACGACGCTGAACCAGATCTGATACCCAATAATTATTGTGAAGCGACGAGCGCGCCTAGTCTCTCCGTGTCGTCTCCCGCGCGGACAGGCCCCGCACCCCTCACCACCCCGGCGTGCAACGGTCCTGGCGCGTCGTGGACGTGGGGCGCGCGGTGCCGGGAGCCGGCGGACGCGGCGCTGGGGCGGAGCGCCTCCGCCACGCTGCGAGCCGCCGCGGACCTCAACGTTGACGCCATCAAAGAGAAACTCTTAGACAATCGAATCCCGGAATCCTGCGTCTAG
Protein
MLETIECLVYMEIEPTTLDAPNFSNSKCCYSFDVRGKPLSEARHWSAPEVFGPRAHFSTSPAPASLLVRDVKRRDEGVYRCRVDFRNTQTTSFRYNLTVVVPPEKPVVVDRWGRVINTTTLGPHEEGDDVLLTCRVLGGRPEPSVRWLVNGVLVDEEYEHNTGDVIENRLLWPAIRRADYAAVFTCQAANSHLVPPKEVSLVLDMFLRPLTVEIKKPAETGEGGVLTAERRYEVACESAGSRPPAIVTWFKGKRQLKRITEELRDNLTMSVMSFVPTLDDDGKPITCRAENPNVTTLSMETSWTINVVYPPVVRLRLGSSLAAGDIKEGDDVYFECHVRANPPARKLSWLHDDRPLAHNASARVFHSNQSLVLQKVTRHSSGRYACSALNAEGETVSNELHFRVKYAPSCRSGGVSVVGAARGESVVIVCKVDADPPAAVFKWKFNNSGETLDVAADRYTSNGSASSLKYTPVADLDYGTLSCSASNEVGAQLAPCVFQMVVAGKPHAPRNCTLWNQTAESVEVSCVAGFDGGLPQTFLLEVYHADGSSPRVNLTSEEPSWTIRGLEWDVRFRLIAVAVNSKGRSPPASLDDVLFRDPEKRTATEASLGMSSMLGAAGAGAGAIFALLAGACAVSLRRRRRPRLAKPLPAPLVTQHLTLKQPELDDAEPDLIPNNYCEATSAPSLSVSSPARTGPAPLTTPACNGPGASWTWGARCREPADAALGRSASATLRAAADLNVDAIKEKLLDNRIPESCV
Summary
Uniprot
A0A139WE49
A0A067QVT8
A0A2Y9D349
A0A2Y9D0W0
A0A1S4H2N4
A0A3F2YUY3
+ More
A0A2Y9D3H7 Q7Q1Q0 A0A336MF21 A0A1I8PYM0 A0A1I8N3L1 B3MLD5 A0A0J9R2I5 B3N5M1 B4MZ69 A0A1W4W7L2 B4P0W6 C4IXX0 B4LRZ2 A0A1J1ILM7 B4KE46 A0A3B0K6G8 Q59DZ1 B4HXK7 A0A1J1IJT1 A0A154P3F0 A0A195EH15 A0A195F6R9 A0A158NIA0 A0A195AWT4 A0A026WW65 A0A182Y5L5 A0A182QB13 A0A151WJS4 Q7Q1W7 A0A336MAP7 B5DH28 A0A1A9ZVX1 A0A1B0B7L8 Q16WY2 A0A0L7R734 W5JDD9 A0A2S2Q9A9 A0A3L8D982 A0A2H8TGU6 A0A2S2NCV8 Q7PN36 A0A1S4GYP1 A0A182XL95 A0A182ICC9 A0A182K5C4 A0A182PTY4 A0A182NTT6 A0A182JAT4 A0A182RMB4 A0A182W6L7 Q16UD9 A0A182F1F4 A0A1A9W080 A0A1W4WFY8 A0A1W4WQT7 A0A1W4WRT9 D6WPK9 A0A2J7PYN3 A0A0K8T359 A0A0A9ZBH5 A0A1Y1NKS5 A0A1B6G259 A0A067RCL4 A0A1I8PBW5 A0A336MBR2 A0A182P2F8 E0VML7 A0A1B6LB71 C0PTV4 Q16ZD1 A0A336M9R3 A0A182X9U7 A0A182Y570 A0A182L937 A0A1I8NIL5 A0A0Q9W1D8 A0A1B6C6V9
A0A2Y9D3H7 Q7Q1Q0 A0A336MF21 A0A1I8PYM0 A0A1I8N3L1 B3MLD5 A0A0J9R2I5 B3N5M1 B4MZ69 A0A1W4W7L2 B4P0W6 C4IXX0 B4LRZ2 A0A1J1ILM7 B4KE46 A0A3B0K6G8 Q59DZ1 B4HXK7 A0A1J1IJT1 A0A154P3F0 A0A195EH15 A0A195F6R9 A0A158NIA0 A0A195AWT4 A0A026WW65 A0A182Y5L5 A0A182QB13 A0A151WJS4 Q7Q1W7 A0A336MAP7 B5DH28 A0A1A9ZVX1 A0A1B0B7L8 Q16WY2 A0A0L7R734 W5JDD9 A0A2S2Q9A9 A0A3L8D982 A0A2H8TGU6 A0A2S2NCV8 Q7PN36 A0A1S4GYP1 A0A182XL95 A0A182ICC9 A0A182K5C4 A0A182PTY4 A0A182NTT6 A0A182JAT4 A0A182RMB4 A0A182W6L7 Q16UD9 A0A182F1F4 A0A1A9W080 A0A1W4WFY8 A0A1W4WQT7 A0A1W4WRT9 D6WPK9 A0A2J7PYN3 A0A0K8T359 A0A0A9ZBH5 A0A1Y1NKS5 A0A1B6G259 A0A067RCL4 A0A1I8PBW5 A0A336MBR2 A0A182P2F8 E0VML7 A0A1B6LB71 C0PTV4 Q16ZD1 A0A336M9R3 A0A182X9U7 A0A182Y570 A0A182L937 A0A1I8NIL5 A0A0Q9W1D8 A0A1B6C6V9
Pubmed
EMBL
KQ971354
KYB26230.1
KK853189
KDR10051.1
APCN01005643
APCN01005644
+ More
APCN01005645 APCN01005646 APCN01005647 AAAB01008980 EAA14139.4 UFQT01001107 SSX28944.1 CH902620 EDV30724.2 CM002910 KMY90306.1 CH954177 EDV57980.2 CH963913 EDW77465.2 CM000157 EDW89040.2 BT083401 BT083402 ACQ83315.1 ACQ83316.1 CH940649 EDW63668.2 CVRI01000055 CRL01064.1 CH933807 EDW11791.2 OUUW01000033 SPP89817.1 AE014134 AAX52666.3 AHN54460.1 CH480818 EDW51787.1 CVRI01000054 CRL00448.1 KQ434809 KZC06465.1 KQ978957 KYN27172.1 KQ981763 KYN36076.1 ADTU01016137 ADTU01016138 ADTU01016139 ADTU01016140 ADTU01016141 ADTU01016142 ADTU01016143 ADTU01016144 ADTU01016145 ADTU01016146 KQ976725 KYM76510.1 KK107078 EZA60257.1 AXCN02000750 KQ983039 KYQ48081.1 EAA14531.4 UFQT01000825 SSX27414.1 CH379058 EDY69751.2 JXJN01009684 JXJN01009685 CH477551 EAT39106.1 KQ414643 KOC66685.1 ADMH02001859 ETN60829.1 GGMS01004947 MBY74150.1 QOIP01000011 RLU16894.1 GFXV01001542 MBW13347.1 GGMR01002432 MBY15051.1 AAAB01008964 EAA12841.5 APCN01000794 AXCP01007831 CH477624 EAT38129.1 EFA06826.2 NEVH01020344 PNF21422.1 GBRD01006221 JAG59600.1 GBHO01002348 GDHC01008300 JAG41256.1 JAQ10329.1 GEZM01002471 JAV97235.1 GECZ01013250 JAS56519.1 KK852551 KDR21492.1 UFQT01000721 SSX26761.1 DS235315 EEB14623.1 GEBQ01019020 JAT20957.1 BT071799 ACN43735.2 CH477492 EAT40016.1 SSX26760.1 CH940652 KRF78738.1 GEDC01028050 JAS09248.1
APCN01005645 APCN01005646 APCN01005647 AAAB01008980 EAA14139.4 UFQT01001107 SSX28944.1 CH902620 EDV30724.2 CM002910 KMY90306.1 CH954177 EDV57980.2 CH963913 EDW77465.2 CM000157 EDW89040.2 BT083401 BT083402 ACQ83315.1 ACQ83316.1 CH940649 EDW63668.2 CVRI01000055 CRL01064.1 CH933807 EDW11791.2 OUUW01000033 SPP89817.1 AE014134 AAX52666.3 AHN54460.1 CH480818 EDW51787.1 CVRI01000054 CRL00448.1 KQ434809 KZC06465.1 KQ978957 KYN27172.1 KQ981763 KYN36076.1 ADTU01016137 ADTU01016138 ADTU01016139 ADTU01016140 ADTU01016141 ADTU01016142 ADTU01016143 ADTU01016144 ADTU01016145 ADTU01016146 KQ976725 KYM76510.1 KK107078 EZA60257.1 AXCN02000750 KQ983039 KYQ48081.1 EAA14531.4 UFQT01000825 SSX27414.1 CH379058 EDY69751.2 JXJN01009684 JXJN01009685 CH477551 EAT39106.1 KQ414643 KOC66685.1 ADMH02001859 ETN60829.1 GGMS01004947 MBY74150.1 QOIP01000011 RLU16894.1 GFXV01001542 MBW13347.1 GGMR01002432 MBY15051.1 AAAB01008964 EAA12841.5 APCN01000794 AXCP01007831 CH477624 EAT38129.1 EFA06826.2 NEVH01020344 PNF21422.1 GBRD01006221 JAG59600.1 GBHO01002348 GDHC01008300 JAG41256.1 JAQ10329.1 GEZM01002471 JAV97235.1 GECZ01013250 JAS56519.1 KK852551 KDR21492.1 UFQT01000721 SSX26761.1 DS235315 EEB14623.1 GEBQ01019020 JAT20957.1 BT071799 ACN43735.2 CH477492 EAT40016.1 SSX26760.1 CH940652 KRF78738.1 GEDC01028050 JAS09248.1
Proteomes
UP000007266
UP000027135
UP000075903
UP000075840
UP000075882
UP000076407
+ More
UP000007062 UP000095300 UP000095301 UP000007801 UP000008711 UP000007798 UP000192221 UP000002282 UP000008792 UP000183832 UP000009192 UP000268350 UP000000803 UP000001292 UP000076502 UP000078492 UP000078541 UP000005205 UP000078540 UP000053097 UP000076408 UP000075886 UP000075809 UP000001819 UP000092445 UP000092460 UP000008820 UP000053825 UP000000673 UP000279307 UP000075881 UP000075885 UP000075884 UP000075880 UP000075900 UP000075920 UP000069272 UP000091820 UP000192223 UP000235965 UP000009046
UP000007062 UP000095300 UP000095301 UP000007801 UP000008711 UP000007798 UP000192221 UP000002282 UP000008792 UP000183832 UP000009192 UP000268350 UP000000803 UP000001292 UP000076502 UP000078492 UP000078541 UP000005205 UP000078540 UP000053097 UP000076408 UP000075886 UP000075809 UP000001819 UP000092445 UP000092460 UP000008820 UP000053825 UP000000673 UP000279307 UP000075881 UP000075885 UP000075884 UP000075880 UP000075900 UP000075920 UP000069272 UP000091820 UP000192223 UP000235965 UP000009046
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A139WE49
A0A067QVT8
A0A2Y9D349
A0A2Y9D0W0
A0A1S4H2N4
A0A3F2YUY3
+ More
A0A2Y9D3H7 Q7Q1Q0 A0A336MF21 A0A1I8PYM0 A0A1I8N3L1 B3MLD5 A0A0J9R2I5 B3N5M1 B4MZ69 A0A1W4W7L2 B4P0W6 C4IXX0 B4LRZ2 A0A1J1ILM7 B4KE46 A0A3B0K6G8 Q59DZ1 B4HXK7 A0A1J1IJT1 A0A154P3F0 A0A195EH15 A0A195F6R9 A0A158NIA0 A0A195AWT4 A0A026WW65 A0A182Y5L5 A0A182QB13 A0A151WJS4 Q7Q1W7 A0A336MAP7 B5DH28 A0A1A9ZVX1 A0A1B0B7L8 Q16WY2 A0A0L7R734 W5JDD9 A0A2S2Q9A9 A0A3L8D982 A0A2H8TGU6 A0A2S2NCV8 Q7PN36 A0A1S4GYP1 A0A182XL95 A0A182ICC9 A0A182K5C4 A0A182PTY4 A0A182NTT6 A0A182JAT4 A0A182RMB4 A0A182W6L7 Q16UD9 A0A182F1F4 A0A1A9W080 A0A1W4WFY8 A0A1W4WQT7 A0A1W4WRT9 D6WPK9 A0A2J7PYN3 A0A0K8T359 A0A0A9ZBH5 A0A1Y1NKS5 A0A1B6G259 A0A067RCL4 A0A1I8PBW5 A0A336MBR2 A0A182P2F8 E0VML7 A0A1B6LB71 C0PTV4 Q16ZD1 A0A336M9R3 A0A182X9U7 A0A182Y570 A0A182L937 A0A1I8NIL5 A0A0Q9W1D8 A0A1B6C6V9
A0A2Y9D3H7 Q7Q1Q0 A0A336MF21 A0A1I8PYM0 A0A1I8N3L1 B3MLD5 A0A0J9R2I5 B3N5M1 B4MZ69 A0A1W4W7L2 B4P0W6 C4IXX0 B4LRZ2 A0A1J1ILM7 B4KE46 A0A3B0K6G8 Q59DZ1 B4HXK7 A0A1J1IJT1 A0A154P3F0 A0A195EH15 A0A195F6R9 A0A158NIA0 A0A195AWT4 A0A026WW65 A0A182Y5L5 A0A182QB13 A0A151WJS4 Q7Q1W7 A0A336MAP7 B5DH28 A0A1A9ZVX1 A0A1B0B7L8 Q16WY2 A0A0L7R734 W5JDD9 A0A2S2Q9A9 A0A3L8D982 A0A2H8TGU6 A0A2S2NCV8 Q7PN36 A0A1S4GYP1 A0A182XL95 A0A182ICC9 A0A182K5C4 A0A182PTY4 A0A182NTT6 A0A182JAT4 A0A182RMB4 A0A182W6L7 Q16UD9 A0A182F1F4 A0A1A9W080 A0A1W4WFY8 A0A1W4WQT7 A0A1W4WRT9 D6WPK9 A0A2J7PYN3 A0A0K8T359 A0A0A9ZBH5 A0A1Y1NKS5 A0A1B6G259 A0A067RCL4 A0A1I8PBW5 A0A336MBR2 A0A182P2F8 E0VML7 A0A1B6LB71 C0PTV4 Q16ZD1 A0A336M9R3 A0A182X9U7 A0A182Y570 A0A182L937 A0A1I8NIL5 A0A0Q9W1D8 A0A1B6C6V9
PDB
6IAA
E-value=2.87736e-14,
Score=194
Ontologies
GO
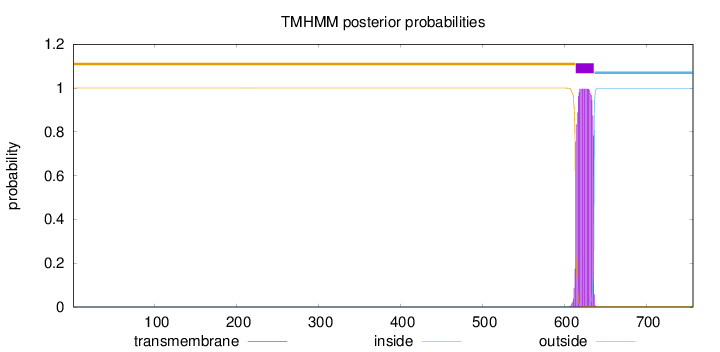
Topology
Length:
757
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.32947
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00004
outside
1 - 613
TMhelix
614 - 636
inside
637 - 757
Population Genetic Test Statistics
Pi
2.510797
Theta
16.413424
Tajima's D
-1.984025
CLR
1785.989943
CSRT
0.0145992700364982
Interpretation
Possibly Positive selection
