Gene
KWMTBOMO02336
Annotation
reverse_transcriptase_[Bombyx_mori]
Full name
Probable RNA-directed DNA polymerase from transposon BS
Alternative Name
Reverse transcriptase
Location in the cell
Mitochondrial Reliability : 1.01 Nuclear Reliability : 1.375
Sequence
CDS
ATGCCCCCCCTCGTAGGCCCCTCAGGCCGACTCGCGGCGTTCGATGATGACGAAAAAGTAGAGCTGCTGGCCGATACATTGCAAGCCCAGTGCACGCCCAGCACTCAATCCGTGGACCCTGTTCATGTAGAATTAGTAGACAGTGAGGTAGAACGCAGAGTCTCCTTGCCACCCTCGGATGCGTTACCACCCGTCACCCCGATGGAAGTTAAAGACTTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCGACGGTATATCCAACCGCGTTATTAAACTTCTACCCGTCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGTATACATAAACCCGGTAAACAAAAAAATCATCCGACGAGCCGCCCGATTAGCCTCCTCATGTCTCTAGGCAAACTGTATGAGCGTCTGCTCTACAAACGCCTCAGAGACTTCGTCTCATCCAAGGGCATTCTCATCGATGAACAATTCGGATTCCGTACAAATCACTCATGCGTTCAACAGGTGCACCGCCTCCCGGAGTACATTCTTGTGGGGCTTAATCGACCAAAACCGTTATACACGGGAGCTCTCTTCTTCGACGTCGCAAAAGCGTTCGACAAAGTCTGGCACAACGGTTTGATTTTCAAACTATTCAACATGGGCGTGCCGAATAGTCTCGTGCTCGTCATAAGGGACTTCTTGTCGAACCGCTCTTTCGATATCGAGTCGAGGGAACCCGCTCCTACCCACGACCTCTCACAGCTGGAGTCCCGCAAGGCTCTGTCCTCTCACTCCTCCTATTTAGCTTATTCGTCAATGATATTCCCGGGTCGCCGCCGACCCAGTTAG
Protein
MPPLVGPSGRLAAFDDDEKVELLADTLQAQCTPSTQSVDPVHVELVDSEVERRVSLPPSDALPPVTPMEVKDLIKDLRPRKAPGSDGISNRVIKLLPVQLIVMLASIFNAAMANCIFPAVWKEADVIGIHKPGKQKNHPTSRPISLLMSLGKLYERLLYKRLRDFVSSKGILIDEQFGFRTNHSCVQQVHRLPEYILVGLNRPKPLYTGALFFDVAKAFDKVWHNGLIFKLFNMGVPNSLVLVIRDFLSNRSFDIESREPAPTHDLSQLESRKALSSHSSYLAYSSMIFPGRRRPS
Summary
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Cofactor
Mg(2+)
Mn(2+)
Mn(2+)
Keywords
Nucleotidyltransferase
RNA-directed DNA polymerase
Transferase
Transposable element
Feature
chain Probable RNA-directed DNA polymerase from transposon BS
Uniprot
Q93137
Q9BPQ0
Q9XXW0
A0A0N1PFV1
O96546
D7EM17
+ More
A0A139W8G9 A0A1Y1NA23 A0A139WA11 A0A2S2Q352 A0A1Y1K0C8 A0A2S2NPT6 A0A139W8K2 A0A2S2PEZ9 K7IN67 A0A087T6I1 J9KLB1 A0A2H8U0H5 A0A1B6J5Z9 V5GNJ2 J9KKL3 A0A2S2QJM7 A0A2S2PBB4 A0A2S2PX53 X1WWC4 V5GNF3 A0A2S2QEE0 X1WXR6 A0A023F6S4 A0A2S2PWA8 A0A023EYS8 A0A2S2RB48 X1WMU5 X1WN18 X1WWX6 A0A2S2QA24 A0A2S2RAN7 A0A2S2PPK8 J9KC83 X1WXE6 A0A2S2R9D4 X1WR00 X1XGU0 A0A087UCW3 A0A2S2N814 J9KEU4 X1WVL6 A0A2S2R4B7 Q95SX7 A0A2L2YN79 X1X2N2 A0A087SWJ5 A0A2S2P698 A0A232ENU1 A0A023F754 A0A2S2QLY2 J9LC15 J9KPF1 R4FPB9 A0A2S2NGV8 X1WXU3 X1WZH0 A0A182GB02 X1WTA1 A0A2S2N6C8 A0A2S2N629 A0A2J7QJ98 X1WMI5 A0A2A4K8E7
A0A139W8G9 A0A1Y1NA23 A0A139WA11 A0A2S2Q352 A0A1Y1K0C8 A0A2S2NPT6 A0A139W8K2 A0A2S2PEZ9 K7IN67 A0A087T6I1 J9KLB1 A0A2H8U0H5 A0A1B6J5Z9 V5GNJ2 J9KKL3 A0A2S2QJM7 A0A2S2PBB4 A0A2S2PX53 X1WWC4 V5GNF3 A0A2S2QEE0 X1WXR6 A0A023F6S4 A0A2S2PWA8 A0A023EYS8 A0A2S2RB48 X1WMU5 X1WN18 X1WWX6 A0A2S2QA24 A0A2S2RAN7 A0A2S2PPK8 J9KC83 X1WXE6 A0A2S2R9D4 X1WR00 X1XGU0 A0A087UCW3 A0A2S2N814 J9KEU4 X1WVL6 A0A2S2R4B7 Q95SX7 A0A2L2YN79 X1X2N2 A0A087SWJ5 A0A2S2P698 A0A232ENU1 A0A023F754 A0A2S2QLY2 J9LC15 J9KPF1 R4FPB9 A0A2S2NGV8 X1WXU3 X1WZH0 A0A182GB02 X1WTA1 A0A2S2N6C8 A0A2S2N629 A0A2J7QJ98 X1WMI5 A0A2A4K8E7
EC Number
2.7.7.49
Pubmed
EMBL
U07847
AAA17752.1
AB055391
BAB21761.1
AB018558
BAA76304.1
+ More
KQ459265 KPJ02064.1 AF081103 AAC72921.1 KQ973412 EFA12510.2 KQ973342 KXZ75569.1 GEZM01008457 JAV94772.1 KQ971729 KYB24751.1 GGMS01002955 MBY72158.1 GEZM01099921 GEZM01099920 GEZM01099918 GEZM01099906 JAV53125.1 GGMR01006581 MBY19200.1 KQ973213 KXZ75606.1 GGMR01015412 MBY28031.1 KK113655 KFM60720.1 ABLF02040174 ABLF02040183 ABLF02055266 GFXV01007093 MBW18898.1 GECU01013097 JAS94609.1 GALX01005299 JAB63167.1 ABLF02029306 ABLF02029314 ABLF02060138 GGMS01008766 MBY77969.1 GGMR01014053 MBY26672.1 GGMS01000737 MBY69940.1 ABLF02003961 ABLF02059872 GALX01005359 JAB63107.1 GGMS01006359 MBY75562.1 ABLF02008605 ABLF02008608 ABLF02008609 ABLF02008610 ABLF02017023 GBBI01001646 JAC17066.1 GGMS01000625 MBY69828.1 GBBI01004395 JAC14317.1 GGMS01018022 MBY87225.1 ABLF02014862 ABLF02014866 ABLF02061908 ABLF02021177 ABLF02027367 ABLF02066338 ABLF02036422 ABLF02042484 GGMS01005381 MBY74584.1 GGMS01017914 MBY87117.1 GGMR01018716 MBY31335.1 ABLF02033757 ABLF02016634 GGMS01017365 MBY86568.1 ABLF02003242 ABLF02006260 ABLF02042516 ABLF02055645 KK119261 KFM75202.1 GGMR01000676 MBY13295.1 ABLF02040650 ABLF02040651 ABLF02056776 ABLF02042385 GGMS01015632 MBY84835.1 AY060438 AAL25477.1 IAAA01035749 LAA09611.1 KK112269 KFM57234.1 GGMR01012305 MBY24924.1 NNAY01003095 OXU19992.1 GBBI01001642 JAC17070.1 GGMS01009480 MBY78683.1 ABLF02020655 ABLF02039168 ABLF02061060 ABLF02041767 GAHY01000238 JAA77272.1 GGMR01003407 MBY16026.1 ABLF02008933 ABLF02034467 JXUM01241784 KQ635293 KXJ62492.1 ABLF02037660 GGMR01000086 MBY12705.1 GGMR01000005 MBY12624.1 NEVH01013558 PNF28668.1 ABLF02004232 ABLF02004856 ABLF02004857 ABLF02035566 ABLF02056910 NWSH01000032 PCG80497.1
KQ459265 KPJ02064.1 AF081103 AAC72921.1 KQ973412 EFA12510.2 KQ973342 KXZ75569.1 GEZM01008457 JAV94772.1 KQ971729 KYB24751.1 GGMS01002955 MBY72158.1 GEZM01099921 GEZM01099920 GEZM01099918 GEZM01099906 JAV53125.1 GGMR01006581 MBY19200.1 KQ973213 KXZ75606.1 GGMR01015412 MBY28031.1 KK113655 KFM60720.1 ABLF02040174 ABLF02040183 ABLF02055266 GFXV01007093 MBW18898.1 GECU01013097 JAS94609.1 GALX01005299 JAB63167.1 ABLF02029306 ABLF02029314 ABLF02060138 GGMS01008766 MBY77969.1 GGMR01014053 MBY26672.1 GGMS01000737 MBY69940.1 ABLF02003961 ABLF02059872 GALX01005359 JAB63107.1 GGMS01006359 MBY75562.1 ABLF02008605 ABLF02008608 ABLF02008609 ABLF02008610 ABLF02017023 GBBI01001646 JAC17066.1 GGMS01000625 MBY69828.1 GBBI01004395 JAC14317.1 GGMS01018022 MBY87225.1 ABLF02014862 ABLF02014866 ABLF02061908 ABLF02021177 ABLF02027367 ABLF02066338 ABLF02036422 ABLF02042484 GGMS01005381 MBY74584.1 GGMS01017914 MBY87117.1 GGMR01018716 MBY31335.1 ABLF02033757 ABLF02016634 GGMS01017365 MBY86568.1 ABLF02003242 ABLF02006260 ABLF02042516 ABLF02055645 KK119261 KFM75202.1 GGMR01000676 MBY13295.1 ABLF02040650 ABLF02040651 ABLF02056776 ABLF02042385 GGMS01015632 MBY84835.1 AY060438 AAL25477.1 IAAA01035749 LAA09611.1 KK112269 KFM57234.1 GGMR01012305 MBY24924.1 NNAY01003095 OXU19992.1 GBBI01001642 JAC17070.1 GGMS01009480 MBY78683.1 ABLF02020655 ABLF02039168 ABLF02061060 ABLF02041767 GAHY01000238 JAA77272.1 GGMR01003407 MBY16026.1 ABLF02008933 ABLF02034467 JXUM01241784 KQ635293 KXJ62492.1 ABLF02037660 GGMR01000086 MBY12705.1 GGMR01000005 MBY12624.1 NEVH01013558 PNF28668.1 ABLF02004232 ABLF02004856 ABLF02004857 ABLF02035566 ABLF02056910 NWSH01000032 PCG80497.1
Proteomes
Pfam
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
Q93137
Q9BPQ0
Q9XXW0
A0A0N1PFV1
O96546
D7EM17
+ More
A0A139W8G9 A0A1Y1NA23 A0A139WA11 A0A2S2Q352 A0A1Y1K0C8 A0A2S2NPT6 A0A139W8K2 A0A2S2PEZ9 K7IN67 A0A087T6I1 J9KLB1 A0A2H8U0H5 A0A1B6J5Z9 V5GNJ2 J9KKL3 A0A2S2QJM7 A0A2S2PBB4 A0A2S2PX53 X1WWC4 V5GNF3 A0A2S2QEE0 X1WXR6 A0A023F6S4 A0A2S2PWA8 A0A023EYS8 A0A2S2RB48 X1WMU5 X1WN18 X1WWX6 A0A2S2QA24 A0A2S2RAN7 A0A2S2PPK8 J9KC83 X1WXE6 A0A2S2R9D4 X1WR00 X1XGU0 A0A087UCW3 A0A2S2N814 J9KEU4 X1WVL6 A0A2S2R4B7 Q95SX7 A0A2L2YN79 X1X2N2 A0A087SWJ5 A0A2S2P698 A0A232ENU1 A0A023F754 A0A2S2QLY2 J9LC15 J9KPF1 R4FPB9 A0A2S2NGV8 X1WXU3 X1WZH0 A0A182GB02 X1WTA1 A0A2S2N6C8 A0A2S2N629 A0A2J7QJ98 X1WMI5 A0A2A4K8E7
A0A139W8G9 A0A1Y1NA23 A0A139WA11 A0A2S2Q352 A0A1Y1K0C8 A0A2S2NPT6 A0A139W8K2 A0A2S2PEZ9 K7IN67 A0A087T6I1 J9KLB1 A0A2H8U0H5 A0A1B6J5Z9 V5GNJ2 J9KKL3 A0A2S2QJM7 A0A2S2PBB4 A0A2S2PX53 X1WWC4 V5GNF3 A0A2S2QEE0 X1WXR6 A0A023F6S4 A0A2S2PWA8 A0A023EYS8 A0A2S2RB48 X1WMU5 X1WN18 X1WWX6 A0A2S2QA24 A0A2S2RAN7 A0A2S2PPK8 J9KC83 X1WXE6 A0A2S2R9D4 X1WR00 X1XGU0 A0A087UCW3 A0A2S2N814 J9KEU4 X1WVL6 A0A2S2R4B7 Q95SX7 A0A2L2YN79 X1X2N2 A0A087SWJ5 A0A2S2P698 A0A232ENU1 A0A023F754 A0A2S2QLY2 J9LC15 J9KPF1 R4FPB9 A0A2S2NGV8 X1WXU3 X1WZH0 A0A182GB02 X1WTA1 A0A2S2N6C8 A0A2S2N629 A0A2J7QJ98 X1WMI5 A0A2A4K8E7
Ontologies
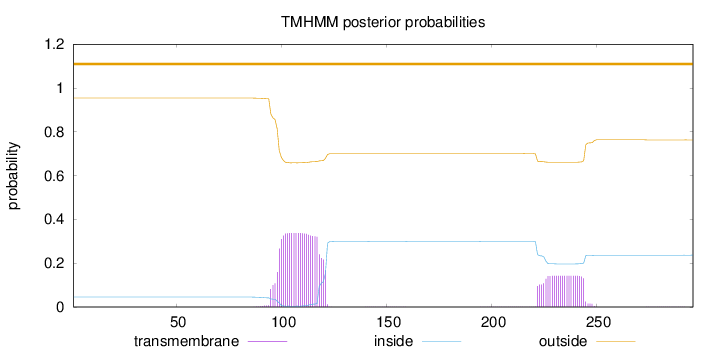
Topology
Length:
296
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.75466
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04537
outside
1 - 296
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
51.790308
CSRT
0
Interpretation
Uncertain
