Gene
KWMTBOMO02332
Annotation
Retrotransposable_element_Tf2_155_kDa_protein_type_1_[Papilio_machaon]
Full name
SAGA-associated factor 11
Location in the cell
Nuclear Reliability : 4.382
Sequence
CDS
ATGGATATTTTTGTGGTGGATAATGAAAATTTTAACTATGACTTCTTAATTGGTTTGGATTGTATAAAAAAATTCAAATTAATACAAAATGAAGAACTTAACATCATACAATATCCCGAAAAGAAAAGCATAATCTCAAATGAACATTGTTCTAATGAGAATGTAACAAATGTCGGAATTCCTGATGTACTAGGTGACAAAAATAAAGAACAAACATTTAAAGAGAGAATAGAAGAGAAAGAGAAGATAGAGATGGAACTATACTCTGAGATAGTAAAAAAGGGTACTACTATAAATACCAACAAATTGTGTGAAATTAATTTCAATGAACATGTAAACATTGATGAATTTCAAATGTCTGTAAATCATCTAGATTTATATCAGCAATCAAAAATTGAGAAGTTAGTAGATAAATACAAATCTGTTTTTGCTAAGGACAAATATGATGTTGGCTCTGTGCAGGATTATGAAGCCCATATAGACTTGATGGTAGATAAATACTGCTATAAGCGTCCGTACAGATGCACAATTGAAGACAGAACAGAAATTGAAAGTCAAATATCGAAGCTACTACAGAAAAATTTGATAGAGGAATCTTACAGCCCGTTTGCTGCTCCTGTAACTCTAGCATATAAGAAAGAAGATGATAAAAAAACAAGATTATGCATAGATTTCAGAGATTTAAACAAAATAGTAGTTCCACAATCACAACCTTTCCCACTTATAGAGGACTTAATGACCAAAACAGTAAACTGTAAGTATTATTCTACATTAGACATAAACTCAGCATTTTGGTCGATCCCATTGAAAATTCAAGATAGACATAAGACTGCTTTTGTGACACAAGAAGGACATTTTCAATGGACTTGTCTCCCATTTGGGTTAAAGACTGCACCAGCGATTTTCCAAAGAATATTGAGCAATATTATAAGAAAACACAGACTATCTAACTTTGCAGTAAACTTTATTGACGACATCTTAATTTTTTCTAAAACTTTTGAGGAACATATACAACACATATCGAGACTACTGGAAGCAATACAAAAGGAAGGGTTTAGACTGAAGTTTTCAAAATGTACATTTGCAAGTCACCATGCTAAATACTTGGGTCATATAATAGAAGACAATTCAGTTAGACCTCTGAAAGATTATTTAACTGCTGTCAAAAGCTTCCCAATTCCAGAAACAAGCAAAAATATACGGCAGTTTTTGGGAAAAGTAAATTTTCATCATAAGTTTGTACCACATAGTGCAATTATCCTCGATCCATTACATAACTTATTAAGGAAAGATGTGAAATTTGTATGGACGGCAGAATGCCAGGAAGCATTTGATAAAATTAGAGCATTGCTTTGCTCAGAACCAATCCTAAAAATATTTGACCCAGATTCACCGATAAGAATTTACACAGATGCTAGTATCAAAGGAGTTGGAGCAGTGTTGAAACAAGACGATGAAAACGGCGTGAGTAAACCGGTTGCTTATTTTTCGAAGAAATTAACAGAAGCACAGAAAAAGAAGAAGGCAATATACTTAGAATGTCTGGCGATAAAAGAAGCAGTCAAATACTGGCAACATTGGCTTATGGGAAAAGAATTTTTGGTATACACAGATCATAAACCCCTGGAAAATATGAACATTAAAGCTCGAACTGATGAGGAACTAGGTGATTTAACATATTATCTATCACAATATAATTTTGAAATTAAATACAACCCAGGAAAATCAAATCAAGAAGCGGACTGCTTAAGCCGAAATCCAGTCCTAGAAGCAAATGATAATAACGACGATTTTTTAAAGGTCGTAAATTTAATCGACCTACAAGAAATAAAGAATGACCAGCTAACAAATCCAGACCTACAAAAAGAAAAGAATAAGCACATATTAGAGAATGAAGTTTATTACAAGAAAAATAAAAGAAGAAATAAGATTATACTGTCAGAAAAACTTAGCAAAGCTCTAATAAAAAAAGTCCATGACACATACTGCCACATTGGAAGAACCCAGATGACAAATAAAATAACACCATTCTTTTCAGCCAAGAATATTACAGCAAACATTAATAAAATGTGTGATGCGTGTGAAACCTGCATTAAAAACAAGTCCAGGAGGAAGTCGAAATATGGTTTAATGTCACATTTGGGACCAGCCACACACCCGTTTGAAATTGTATCCATCGATACTATTGGGGGCTTTGGAGGATCCCGATCAACCAAAAGAGAAGGGTATAAATTGGAAGAAGGAGATAGTGTTTATGTGGAGAATGGAAATAAACTAAACAGGAAGAAAATGGATGAATTGAGAACAGGGCCATACAAAATACTAAAGAAGATTTCAGAATCGATCTATGAAGTTGACACCGGACACAAAAAAGCAGAATCAAACCTCTATCATATAACAAAGTTGTCTCCAGTGTCTATCTGA
Protein
MDIFVVDNENFNYDFLIGLDCIKKFKLIQNEELNIIQYPEKKSIISNEHCSNENVTNVGIPDVLGDKNKEQTFKERIEEKEKIEMELYSEIVKKGTTINTNKLCEINFNEHVNIDEFQMSVNHLDLYQQSKIEKLVDKYKSVFAKDKYDVGSVQDYEAHIDLMVDKYCYKRPYRCTIEDRTEIESQISKLLQKNLIEESYSPFAAPVTLAYKKEDDKKTRLCIDFRDLNKIVVPQSQPFPLIEDLMTKTVNCKYYSTLDINSAFWSIPLKIQDRHKTAFVTQEGHFQWTCLPFGLKTAPAIFQRILSNIIRKHRLSNFAVNFIDDILIFSKTFEEHIQHISRLLEAIQKEGFRLKFSKCTFASHHAKYLGHIIEDNSVRPLKDYLTAVKSFPIPETSKNIRQFLGKVNFHHKFVPHSAIILDPLHNLLRKDVKFVWTAECQEAFDKIRALLCSEPILKIFDPDSPIRIYTDASIKGVGAVLKQDDENGVSKPVAYFSKKLTEAQKKKKAIYLECLAIKEAVKYWQHWLMGKEFLVYTDHKPLENMNIKARTDEELGDLTYYLSQYNFEIKYNPGKSNQEADCLSRNPVLEANDNNDDFLKVVNLIDLQEIKNDQLTNPDLQKEKNKHILENEVYYKKNKRRNKIILSEKLSKALIKKVHDTYCHIGRTQMTNKITPFFSAKNITANINKMCDACETCIKNKSRRKSKYGLMSHLGPATHPFEIVSIDTIGGFGGSRSTKREGYKLEEGDSVYVENGNKLNRKKMDELRTGPYKILKKISESIYEVDTGHKKAESNLYHITKLSPVSI
Summary
Similarity
Belongs to the SGF11 family.
Belongs to the tubulin family.
Belongs to the tubulin family.
Uniprot
A0A0N1PG61
A0A2S2NJQ2
A0A0K8TMU5
A0A0N0PCL2
A0A0J7K739
A0A1B6C1P7
+ More
A0A1B6DVG3 A0A1B6E784 A0A0K8TLR3 A0A2S2PQS7 A0A2R7W771 A0A2R7WSP7 A0A1B6CSL2 A0A2L2Y3Y7 A0A2L2Y4F2 A0A087U2Q4 A0A158P4Q1 A0A158P4S6 A0A158P5I9 L7MCS8 L7MA76 A0A0C9RXL6 A0A388K567 A0A090XDG5 A0A1E1XL52 L7LW12 L7LW11 L7LYQ9 A0A388KWR7 A0A1E1XKD7 L7LV95 L7ML53 L7LY95 A0A1E1X293 A0A388LMS8 L7LYP4 A0A388MA01 A0A147BMC5 L7MFA7 A0A388JJI1 A0A388KXH3 A0A1Y1KJI2 L7MAW1 A0A388KWG6 L7MBZ6 L7LV55 A0A388K9A8 A0A388KQG1 A0A388L150 A0A388LDS5 A0A388LN87 A0A388KQM9 L7MML3 L7LVQ9 A0A2R5L8F2 A0A388JUK0 A0A388KZE6 A0A388L2E3 A0A388M7C9 A0A2R5LEZ4 A0A388KIW5 A0A1Y1KFN9 A0A1Y1KFM9 A0A2R5LP11 A0A147BJI2 L7ML73 A0A388KSU5 A0A388L5S1 A0A034VPR8 A0A147BAZ1 A0A139WIQ0 L7LWL7 A0A131XNQ4 A0A139WJA1 A0A147BIK4 A0A388K1C8 A0A388JV64 A0A355ABF2
A0A1B6DVG3 A0A1B6E784 A0A0K8TLR3 A0A2S2PQS7 A0A2R7W771 A0A2R7WSP7 A0A1B6CSL2 A0A2L2Y3Y7 A0A2L2Y4F2 A0A087U2Q4 A0A158P4Q1 A0A158P4S6 A0A158P5I9 L7MCS8 L7MA76 A0A0C9RXL6 A0A388K567 A0A090XDG5 A0A1E1XL52 L7LW12 L7LW11 L7LYQ9 A0A388KWR7 A0A1E1XKD7 L7LV95 L7ML53 L7LY95 A0A1E1X293 A0A388LMS8 L7LYP4 A0A388MA01 A0A147BMC5 L7MFA7 A0A388JJI1 A0A388KXH3 A0A1Y1KJI2 L7MAW1 A0A388KWG6 L7MBZ6 L7LV55 A0A388K9A8 A0A388KQG1 A0A388L150 A0A388LDS5 A0A388LN87 A0A388KQM9 L7MML3 L7LVQ9 A0A2R5L8F2 A0A388JUK0 A0A388KZE6 A0A388L2E3 A0A388M7C9 A0A2R5LEZ4 A0A388KIW5 A0A1Y1KFN9 A0A1Y1KFM9 A0A2R5LP11 A0A147BJI2 L7ML73 A0A388KSU5 A0A388L5S1 A0A034VPR8 A0A147BAZ1 A0A139WIQ0 L7LWL7 A0A131XNQ4 A0A139WJA1 A0A147BIK4 A0A388K1C8 A0A388JV64 A0A355ABF2
Pubmed
EMBL
KQ460869
KPJ11799.1
GGMR01004768
MBY17387.1
GDAI01002358
JAI15245.1
+ More
KQ460501 KPJ14174.1 LBMM01012506 KMQ86187.1 GEDC01029866 JAS07432.1 GEDC01007645 JAS29653.1 GEDC01003500 JAS33798.1 GDAI01002319 JAI15284.1 GGMR01019146 MBY31765.1 KK854406 PTY15552.1 KK855443 PTY22588.1 GEDC01020941 JAS16357.1 IAAA01005657 LAA02892.1 IAAA01005656 LAA02887.1 KK117876 KFM71643.1 CAEY01001805 CAEY01001938 CAEY01000944 GACK01004001 JAA61033.1 GACK01004074 JAA60960.1 GBYB01013920 JAG83687.1 BFEA01000059 GBG65156.1 GBIH01000760 JAC93950.1 GFAA01003618 JAT99816.1 GACK01009237 JAA55797.1 GACK01009039 JAA55995.1 GACK01009040 JAA55994.1 BFEA01000202 GBG74403.1 GFAA01003652 JAT99782.1 GACK01009043 JAA55991.1 GACK01000334 JAA64700.1 GACK01009195 JAA55839.1 GFAC01005811 JAT93377.1 BFEA01000446 GBG83646.1 GACK01009041 JAA55993.1 BFEA01000891 GBG91289.1 GEGO01003466 JAR91938.1 GACK01003086 JAA61948.1 BFEA01002572 GBG41953.1 BFEA01000209 GBG74741.1 GEZM01081635 JAV61573.1 GACK01004027 JAA61007.1 GBG74399.1 GACK01003662 JAA61372.1 GACK01009547 JAA55487.1 BFEA01000076 GBG66617.1 BFEA01000162 GBG72311.1 BFEA01000235 GBG75978.1 BFEA01000347 GBG80460.1 BFEA01000451 GBG83778.1 BFEA01000163 GBG72342.1 GACK01000375 JAA64659.1 GACK01009357 JAA55677.1 GGLE01001609 MBY05735.1 BFEA01000020 GBG61486.1 BFEA01000222 GBG75323.1 BFEA01000245 GBG76456.1 BFEA01000809 GBG90415.1 GGLE01003869 MBY07995.1 BFEA01000123 GBG70001.1 GEZM01089268 GEZM01089261 JAV57577.1 GEZM01089269 GEZM01089265 JAV57567.1 GGLE01007069 MBY11195.1 GEGO01004488 JAR90916.1 GACK01000487 JAA64547.1 BFEA01000178 GBG73141.1 BFEA01000273 GBG77650.1 GAKP01014483 JAC44469.1 GEGO01007454 JAR87950.1 KQ971338 KYB27888.1 GACK01009042 JAA55992.1 GEFM01006973 JAP68823.1 KYB27887.1 GEGO01004785 JAR90619.1 BFEA01000043 GBG63850.1 BFEA01000021 GBG61643.1 DNYW01000170 HBK71150.1
KQ460501 KPJ14174.1 LBMM01012506 KMQ86187.1 GEDC01029866 JAS07432.1 GEDC01007645 JAS29653.1 GEDC01003500 JAS33798.1 GDAI01002319 JAI15284.1 GGMR01019146 MBY31765.1 KK854406 PTY15552.1 KK855443 PTY22588.1 GEDC01020941 JAS16357.1 IAAA01005657 LAA02892.1 IAAA01005656 LAA02887.1 KK117876 KFM71643.1 CAEY01001805 CAEY01001938 CAEY01000944 GACK01004001 JAA61033.1 GACK01004074 JAA60960.1 GBYB01013920 JAG83687.1 BFEA01000059 GBG65156.1 GBIH01000760 JAC93950.1 GFAA01003618 JAT99816.1 GACK01009237 JAA55797.1 GACK01009039 JAA55995.1 GACK01009040 JAA55994.1 BFEA01000202 GBG74403.1 GFAA01003652 JAT99782.1 GACK01009043 JAA55991.1 GACK01000334 JAA64700.1 GACK01009195 JAA55839.1 GFAC01005811 JAT93377.1 BFEA01000446 GBG83646.1 GACK01009041 JAA55993.1 BFEA01000891 GBG91289.1 GEGO01003466 JAR91938.1 GACK01003086 JAA61948.1 BFEA01002572 GBG41953.1 BFEA01000209 GBG74741.1 GEZM01081635 JAV61573.1 GACK01004027 JAA61007.1 GBG74399.1 GACK01003662 JAA61372.1 GACK01009547 JAA55487.1 BFEA01000076 GBG66617.1 BFEA01000162 GBG72311.1 BFEA01000235 GBG75978.1 BFEA01000347 GBG80460.1 BFEA01000451 GBG83778.1 BFEA01000163 GBG72342.1 GACK01000375 JAA64659.1 GACK01009357 JAA55677.1 GGLE01001609 MBY05735.1 BFEA01000020 GBG61486.1 BFEA01000222 GBG75323.1 BFEA01000245 GBG76456.1 BFEA01000809 GBG90415.1 GGLE01003869 MBY07995.1 BFEA01000123 GBG70001.1 GEZM01089268 GEZM01089261 JAV57577.1 GEZM01089269 GEZM01089265 JAV57567.1 GGLE01007069 MBY11195.1 GEGO01004488 JAR90916.1 GACK01000487 JAA64547.1 BFEA01000178 GBG73141.1 BFEA01000273 GBG77650.1 GAKP01014483 JAC44469.1 GEGO01007454 JAR87950.1 KQ971338 KYB27888.1 GACK01009042 JAA55992.1 GEFM01006973 JAP68823.1 KYB27887.1 GEGO01004785 JAR90619.1 BFEA01000043 GBG63850.1 BFEA01000021 GBG61643.1 DNYW01000170 HBK71150.1
Proteomes
PRIDE
Pfam
Interpro
IPR041588
Integrase_H2C2
+ More
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR012337 RNaseH-like_sf
IPR041373 RT_RNaseH
IPR002492 Transposase_Tc1-like
IPR038717 Tc1-like_DDE_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR013246 SAGA_su_Sgf11
IPR019103 Peptidase_aspartic_DDI1-type
IPR001878 Znf_CCHC
IPR016197 Chromo-like_dom_sf
IPR001995 Peptidase_A2_cat
IPR036875 Znf_CCHC_sf
IPR025836 Zn_knuckle_CX2CX4HX4C
IPR034122 Retropepsin-like_bacterial
IPR017975 Tubulin_CS
IPR037103 Tubulin/FtsZ_C_sf
IPR003008 Tubulin_FtsZ_GTPase
IPR008280 Tub_FtsZ_C
IPR000217 Tubulin
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR023123 Tubulin_C
IPR002454 Gamma_tubulin
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR018061 Retropepsins
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR012337 RNaseH-like_sf
IPR041373 RT_RNaseH
IPR002492 Transposase_Tc1-like
IPR038717 Tc1-like_DDE_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR013246 SAGA_su_Sgf11
IPR019103 Peptidase_aspartic_DDI1-type
IPR001878 Znf_CCHC
IPR016197 Chromo-like_dom_sf
IPR001995 Peptidase_A2_cat
IPR036875 Znf_CCHC_sf
IPR025836 Zn_knuckle_CX2CX4HX4C
IPR034122 Retropepsin-like_bacterial
IPR017975 Tubulin_CS
IPR037103 Tubulin/FtsZ_C_sf
IPR003008 Tubulin_FtsZ_GTPase
IPR008280 Tub_FtsZ_C
IPR000217 Tubulin
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR023123 Tubulin_C
IPR002454 Gamma_tubulin
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR018061 Retropepsins
SUPFAM
ProteinModelPortal
A0A0N1PG61
A0A2S2NJQ2
A0A0K8TMU5
A0A0N0PCL2
A0A0J7K739
A0A1B6C1P7
+ More
A0A1B6DVG3 A0A1B6E784 A0A0K8TLR3 A0A2S2PQS7 A0A2R7W771 A0A2R7WSP7 A0A1B6CSL2 A0A2L2Y3Y7 A0A2L2Y4F2 A0A087U2Q4 A0A158P4Q1 A0A158P4S6 A0A158P5I9 L7MCS8 L7MA76 A0A0C9RXL6 A0A388K567 A0A090XDG5 A0A1E1XL52 L7LW12 L7LW11 L7LYQ9 A0A388KWR7 A0A1E1XKD7 L7LV95 L7ML53 L7LY95 A0A1E1X293 A0A388LMS8 L7LYP4 A0A388MA01 A0A147BMC5 L7MFA7 A0A388JJI1 A0A388KXH3 A0A1Y1KJI2 L7MAW1 A0A388KWG6 L7MBZ6 L7LV55 A0A388K9A8 A0A388KQG1 A0A388L150 A0A388LDS5 A0A388LN87 A0A388KQM9 L7MML3 L7LVQ9 A0A2R5L8F2 A0A388JUK0 A0A388KZE6 A0A388L2E3 A0A388M7C9 A0A2R5LEZ4 A0A388KIW5 A0A1Y1KFN9 A0A1Y1KFM9 A0A2R5LP11 A0A147BJI2 L7ML73 A0A388KSU5 A0A388L5S1 A0A034VPR8 A0A147BAZ1 A0A139WIQ0 L7LWL7 A0A131XNQ4 A0A139WJA1 A0A147BIK4 A0A388K1C8 A0A388JV64 A0A355ABF2
A0A1B6DVG3 A0A1B6E784 A0A0K8TLR3 A0A2S2PQS7 A0A2R7W771 A0A2R7WSP7 A0A1B6CSL2 A0A2L2Y3Y7 A0A2L2Y4F2 A0A087U2Q4 A0A158P4Q1 A0A158P4S6 A0A158P5I9 L7MCS8 L7MA76 A0A0C9RXL6 A0A388K567 A0A090XDG5 A0A1E1XL52 L7LW12 L7LW11 L7LYQ9 A0A388KWR7 A0A1E1XKD7 L7LV95 L7ML53 L7LY95 A0A1E1X293 A0A388LMS8 L7LYP4 A0A388MA01 A0A147BMC5 L7MFA7 A0A388JJI1 A0A388KXH3 A0A1Y1KJI2 L7MAW1 A0A388KWG6 L7MBZ6 L7LV55 A0A388K9A8 A0A388KQG1 A0A388L150 A0A388LDS5 A0A388LN87 A0A388KQM9 L7MML3 L7LVQ9 A0A2R5L8F2 A0A388JUK0 A0A388KZE6 A0A388L2E3 A0A388M7C9 A0A2R5LEZ4 A0A388KIW5 A0A1Y1KFN9 A0A1Y1KFM9 A0A2R5LP11 A0A147BJI2 L7ML73 A0A388KSU5 A0A388L5S1 A0A034VPR8 A0A147BAZ1 A0A139WIQ0 L7LWL7 A0A131XNQ4 A0A139WJA1 A0A147BIK4 A0A388K1C8 A0A388JV64 A0A355ABF2
PDB
4OL8
E-value=1.46448e-70,
Score=680
Ontologies
GO
PANTHER
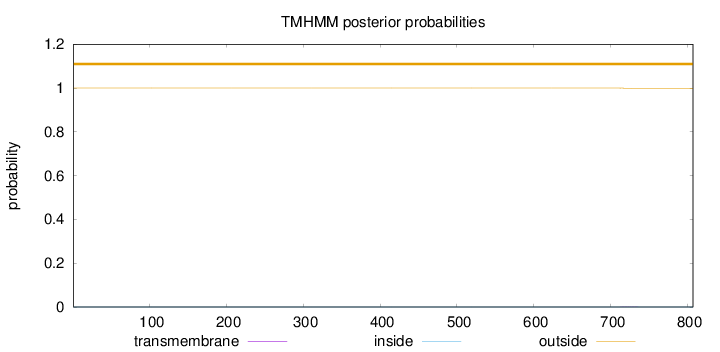
Topology
Subcellular location
Nucleus
Length:
807
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0095
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00011
outside
1 - 807
Population Genetic Test Statistics
Pi
13.168701
Theta
23.374272
Tajima's D
-1.03803
CLR
57.554658
CSRT
0.131293435328234
Interpretation
Uncertain
