Pre Gene Modal
BGIBMGA003759
Annotation
PREDICTED:_transcription_elongation_factor_SPT5_isoform_X1_[Papilio_machaon]
Full name
Transcription elongation factor SPT5
+ More
S-adenosylmethionine sensor upstream of mTORC1
S-adenosylmethionine sensor upstream of mTORC1
Alternative Name
Probable methyltransferase BMT2 homolog
DRB sensitivity-inducing factor large subunit
dSpt5
DRB sensitivity-inducing factor large subunit
dSpt5
Location in the cell
Nuclear Reliability : 2.692
Sequence
CDS
ATGTCGGATTCGGAAGGTAGTAACTATTCGGGGAGTGGGTCCGAGGCAGGCAGTGCTGTATCAAAACAATCACATCGCAGCGCTGCATCGAATCGGTCGGCAAGATCGCGTTCTATCTCTCGATCAAGGTCCAGGTCGCGTTCTGGAAGCCGTAGCCCCTCTAGGACTCCATCCCGCTCACGATCTAGGTCTCGCTCTAGAAGTCGTTCAGTAGGTTCAGATGCAAGTAAAAATAGAGATGATGAAGCCAAAGAAGCATCTGGCGATGAAGAAGTGGAGGATGAACAAGAGCCCGAAGGTGAGGATTTGGTTGATTCTGAGGAGTATGAAGAAGACGAAGAAGAGGAGAGGCTGAGGAAGAAAAGAAAAAAGGACAGTCGTTATGGTGGATTCATTATTGACGAAGCTGAGGTGGATGATGAAGTTGATGAAGATGACGAATGGGAAGAGGGAGCCCAAGAAATGGGTATTGTAGGAAATGAAGTAGATGAAATTGGGCCAACAGCTAGAGAAATTGAAGGTCGAAGAAGAGGCACTAACCTTTGGGATTCACAGAAAGAGGAGGAGATTGAAGAATATCTTCGAAATAAGTATGCAGATGAATCAGCAGCCCTCAGACACTTTGGTGAAGGTGGAGAAGAGATGTCTGATGAAATAACTCAGCAAACTCTACTACCCGGTATCAAGGATCCAAATTTGTGGATGGTCAGATGTAGGATTGGAGAAGAGAAGGCTACTGTTTTACTTTTGATGCGAAAATTTATTGCTTTCCAATTCTCGGAAGAACCTTTTTTGATAAAGTCAGTTGTAGCACCAGAAGGAGTCAAAGGCTATGTGTACATAGAGGCTTTCAAGCAGACACATGTTAAAAACATTATAGACAAAGTGGGAACCCTTAAGTCTGGAGTGTGGAAACAAGAAATGGTGCCTATTAAGGAAATGACTGATGTTCTCAGAGTAGTCAAAAAGCAATCAGGACTTAAACCGAAACAATGGGTCAGATTAAAACGAGGTTTATACAAAGATGATATTGCTCAAGTTGACTATGTAGACTTGGCACAGAATCAAGTGCATTTGAAACTACTTCCTAGAATAGATTATACAAGACTGAGAGGAGCTTTAAGAACAGTACAGAGCGATAGTGAAGCAGCAAAGCGAAAGAAAAAGAGAAGGCCTCCTGCTAAACCCTTTGATCCAGAGGCAATCCGAGCGATCGGTGGAGAAGTTACTTCCGATGGTGATTTCCTCATATTTGAGGGCAATAGGTACTCTAGGAAAGGATTTCTGTATAAAAACTTTACAATGTCGGCGATACTGTCAGAAGGTGTCAAGCCTTCACTTACAGAGTTGGAAAGATTTGAAGAGCAACCGGAAGGTATCGATATTGAGTTGGCTGCTCCCGCCAAGGACGACCCTGCCGGCTTGCATTCCTTCTCCATGGGTGACAATGTGGAAGTCTGTACTGGAGATCTTGCGAACTTGCAGGCCCGCATCACTGCTATCGACGGTTCGATGATCACTGTCATGCCAAAACACGATGGTCTTAAGGACCCAATTGTCTTCCATCCGAATGAGTTGCGCAAGTACTTCAAAATGGGTGATCACGTGAAAGTGGTGGCTGGGCGTTACGAGGGCGACACAGGCCTCATAGTGCGCGTCGAACCGCACAGGGTCGTGCTTGTTTCCGACCTCACCATGCACGAGCTTGAGGTGCTGCCCCGTGACCTACAGCTCTGTTCAGACATGGCGACTGGCGTTGACTCCCTCGGACAGTTCCAGTGGGGAGACATGGTGCAACTGGATCCACAGACCGTCGGAGTTATCGTGCGCCTCGAGAAAGAGAACTTCCACGTGCTCGGCATGCAGGGCAAGGTCATCGAGTGCAAGCCGCAAGCGCTACAGAAACGCCGCGAGAACCGGTTTACAGTCGCGCTCGATTCCGAGGAGAACACGATTCAGAAGCGCGACATCGTTAAAGTAATCGACGGGCCGCATGCCGGACGCGACGGTGAAATCAAACATCTCTACCGGAACTTCGCCTTCCTCCAGTCGCACATGTACCTCGACAACGGCGGCATCTTCGTGTGCAAGACGCGCCACCTGCAGCTGGCCGGTGGAGCCAGGGGAAATAATGCCACCGCCGCCGGACTCACGCTCGGCTTCATGTCGCCGCGGATAGCTTCTCCTGCACACCCAGGGGCCGGCAGGGGAGGCTCCACCCCTCGCGGTCGTGGGGGCCGAGGCGGCGGCGGAGGTCGAGGCGTCACTCGGGATCGCGAGCTCATTGGCCAAACCATTAAGATCACGGGGGGACCATACAAGGGAAACGTCGGGATCGTCAAAGACGCGACCGGCAGCACCGCGCGCGTCGAGCTGCACTCCTCCTGTCAGACCATCTCCGTGGACCGCGGGCACATCGCCGGGGCGGGGGCCGGGGCTGCGACCGGCGGGGCCTCGTCCTACTCGCGCACGCCGTCCCGCACGCCGGCCGCCGGCGCCGCCACCCCCACGTACCGCGACACGGGCATCAAGACGCCTATGCACGGCGCCGCTACACCCGTCTACGAGGTGGGCTCGCGCACGCCACACTACGGGTCGTCGACCCCGGCGCACGAGGGGTCGCGCACGCCGGCGCACGGGGCCTGGGACCCGAGCGGTGCCGCCACCCCGGCGCGACCGGACTTCGACTACGAGCCGGACGCGTACGGCGCGGGACCGTTCACTCCGCAGACGCCCGGCGCCATGTACGGCTCGGACCACACGTACAGCCCGTACAGTCCGGGCCGGTCGGAGGGCGAGTCGGCGGAGGCGGGTGCGTGGGCGGCACCAGAGCTCGTGGTGCGTCTGAGAGCGGGAGCTGCGGACGGTCTGGCCGCACAGACCGCGGTGGTCCGTGCGACCGGCGGGGGCTCGTGCGCGTTGTACTTACCGGCGGAGGAGCGAGTGGTGTCCGTGCCGCCGGACTTGTTAGAGCCGGTGCTGCCTCAGGCGGGAGAGCGCGTGAAGGTGATCGCGGGAGAGGACCGGGACGCGGTCGGACAACTTATCTCCATCGAAAACCAAGAGGGAGTCGTCAAATTCGGCACCGACGACATCAAAATCATGCAACTACGTCATCTTTGCAAGATGGCCTCTTAA
Protein
MSDSEGSNYSGSGSEAGSAVSKQSHRSAASNRSARSRSISRSRSRSRSGSRSPSRTPSRSRSRSRSRSRSVGSDASKNRDDEAKEASGDEEVEDEQEPEGEDLVDSEEYEEDEEEERLRKKRKKDSRYGGFIIDEAEVDDEVDEDDEWEEGAQEMGIVGNEVDEIGPTAREIEGRRRGTNLWDSQKEEEIEEYLRNKYADESAALRHFGEGGEEMSDEITQQTLLPGIKDPNLWMVRCRIGEEKATVLLLMRKFIAFQFSEEPFLIKSVVAPEGVKGYVYIEAFKQTHVKNIIDKVGTLKSGVWKQEMVPIKEMTDVLRVVKKQSGLKPKQWVRLKRGLYKDDIAQVDYVDLAQNQVHLKLLPRIDYTRLRGALRTVQSDSEAAKRKKKRRPPAKPFDPEAIRAIGGEVTSDGDFLIFEGNRYSRKGFLYKNFTMSAILSEGVKPSLTELERFEEQPEGIDIELAAPAKDDPAGLHSFSMGDNVEVCTGDLANLQARITAIDGSMITVMPKHDGLKDPIVFHPNELRKYFKMGDHVKVVAGRYEGDTGLIVRVEPHRVVLVSDLTMHELEVLPRDLQLCSDMATGVDSLGQFQWGDMVQLDPQTVGVIVRLEKENFHVLGMQGKVIECKPQALQKRRENRFTVALDSEENTIQKRDIVKVIDGPHAGRDGEIKHLYRNFAFLQSHMYLDNGGIFVCKTRHLQLAGGARGNNATAAGLTLGFMSPRIASPAHPGAGRGGSTPRGRGGRGGGGGRGVTRDRELIGQTIKITGGPYKGNVGIVKDATGSTARVELHSSCQTISVDRGHIAGAGAGAATGGASSYSRTPSRTPAAGAATPTYRDTGIKTPMHGAATPVYEVGSRTPHYGSSTPAHEGSRTPAHGAWDPSGAATPARPDFDYEPDAYGAGPFTPQTPGAMYGSDHTYSPYSPGRSEGESAEAGAWAAPELVVRLRAGAADGLAAQTAVVRATGGGSCALYLPAEERVVSVPPDLLEPVLPQAGERVKVIAGEDRDAVGQLISIENQEGVVKFGTDDIKIMQLRHLCKMAS
Summary
Description
S-adenosyl-L-methionine-binding protein that acts as an inhibitor of mTORC1 signaling. Acts as a sensor of S-adenosyl-L-methionine to signal methionine sufficiency to mTORC1. Probably also acts as a S-adenosyl-L-methionine-dependent methyltransferase.
Component of the DRB sensitivity-inducing factor complex (DSIF complex), which regulates transcription elongation by RNA polymerase II. DSIF enhances transcriptional pausing at sites proximal to the promoter, which may facilitate the assembly of an elongation competent RNA polymerase II complex. DSIF may also promote transcriptional elongation within coding regions. DSIF is required for the transcriptional induction of heat shock response genes and regulation of genes which control anterior-posterior patterning during embryonic development.
Component of the DRB sensitivity-inducing factor complex (DSIF complex), which regulates transcription elongation by RNA polymerase II. DSIF enhances transcriptional pausing at sites proximal to the promoter, which may facilitate the assembly of an elongation competent RNA polymerase II complex. DSIF may also promote transcriptional elongation within coding regions. DSIF is required for the transcriptional induction of heat shock response genes and regulation of genes which control anterior-posterior patterning during embryonic development.
Subunit
Interacts with Spt6. Interacts with Spt4 to form DSIF. DSIF interacts with trx, RNA polymerase II and with the FACT complex, which is composed of dre4/Spt16 and Ssrp/Ssrp1. DSIF can also interact with the exosome, a complex with 3'-5' exoribonuclease activity which is composed of at least Csl4, Dis3, Mtr3, Rrp4, Rrp6, Rrp40, Rrp42, Rrp46 and Ski6. DSIF may also interact with the positive transcription elongation factor b complex (P-TEFb complex), which is composed of Cdk9 and cyclin-T (CycT).
Similarity
Belongs to the SPT5 family.
Belongs to the BMT2 family.
Belongs to the BMT2 family.
Keywords
Activator
Alternative splicing
Chromosome
Complete proteome
Nucleus
Phosphoprotein
Reference proteome
Repeat
Repressor
Transcription
Transcription regulation
Feature
chain Transcription elongation factor SPT5
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9J2M2
A0A2H1V6P2
A0A194QP01
A0A194QGZ7
A0A212EH19
A0A2A4K3K3
+ More
K7J5S6 A0A1S4FZD5 A0A0P6IVB5 E2B534 A0A182G1W4 A0A182G3H6 A0A1I8NZL8 A0A195CEG6 A0A154P7Y4 E2A5T8 A0A0M9A996 A0A158NYZ9 A0A026W9W4 F4W9U3 A0A232EKF0 E9IQE3 A0A151WKL0 A0A195BQD0 A0A1B0EUS0 A0A182PF58 A0A084WDR7 A0A336MSR5 A0A182K2J7 U5EVY9 A0A182U6J2 A0A182KYR8 A0NE83 A0A182HQS7 A0A182XJE1 A0A182VIZ1 A0A182RUU0 A0A182M7H4 A0A182YLF7 A0A182N1S7 B4PB84 A0A232EM50 A0A0L7R2I7 A0A182QPP4 A0A182JHT9 A0A1S4J862 B3NJV0 A0A1Q3F0W5 B4MRQ6 A0A182WM74 B4LNW9 B4KME4 B0W6Z7 A0A2M3Z117 A0A182FLB7 W5J4B3 A0A0M3QVL5 Q9V460 W8C3N1 B4HPG3 A0A0J9RGS1 A0A2M4BBB8 A0A1W4VGR7 A0A1J1IGM6 Q292S1 B4GCQ3 A0A3B0J2C6 B3MGG2 A0A2M4BBF7 A0A2M4A8G2 B4J626 A0A034WJE4 A0A0L0CNG6 A0A1B0D500 D2A605 A0A1A9WAC5 A0A1B0FL32 A0A1A9YJP3 A0A1A9UYD5 A0A1B0ARK9 A0A0C9PNW0 A0A0A1WN47 A0A1A9ZRD1 A0A1I8M549 A0A2J7Q156 U4UN14 A0A088A942 E0VS63 A0A2A3E981 A0A0C9QH50 A0A023F4S8 A0A0V0G4H6 A0A069DXS5 A0A0P4VJ54 A0A224XHF9 T1I785 A0A2H8TMI3 A0A2S2Q4T6
K7J5S6 A0A1S4FZD5 A0A0P6IVB5 E2B534 A0A182G1W4 A0A182G3H6 A0A1I8NZL8 A0A195CEG6 A0A154P7Y4 E2A5T8 A0A0M9A996 A0A158NYZ9 A0A026W9W4 F4W9U3 A0A232EKF0 E9IQE3 A0A151WKL0 A0A195BQD0 A0A1B0EUS0 A0A182PF58 A0A084WDR7 A0A336MSR5 A0A182K2J7 U5EVY9 A0A182U6J2 A0A182KYR8 A0NE83 A0A182HQS7 A0A182XJE1 A0A182VIZ1 A0A182RUU0 A0A182M7H4 A0A182YLF7 A0A182N1S7 B4PB84 A0A232EM50 A0A0L7R2I7 A0A182QPP4 A0A182JHT9 A0A1S4J862 B3NJV0 A0A1Q3F0W5 B4MRQ6 A0A182WM74 B4LNW9 B4KME4 B0W6Z7 A0A2M3Z117 A0A182FLB7 W5J4B3 A0A0M3QVL5 Q9V460 W8C3N1 B4HPG3 A0A0J9RGS1 A0A2M4BBB8 A0A1W4VGR7 A0A1J1IGM6 Q292S1 B4GCQ3 A0A3B0J2C6 B3MGG2 A0A2M4BBF7 A0A2M4A8G2 B4J626 A0A034WJE4 A0A0L0CNG6 A0A1B0D500 D2A605 A0A1A9WAC5 A0A1B0FL32 A0A1A9YJP3 A0A1A9UYD5 A0A1B0ARK9 A0A0C9PNW0 A0A0A1WN47 A0A1A9ZRD1 A0A1I8M549 A0A2J7Q156 U4UN14 A0A088A942 E0VS63 A0A2A3E981 A0A0C9QH50 A0A023F4S8 A0A0V0G4H6 A0A069DXS5 A0A0P4VJ54 A0A224XHF9 T1I785 A0A2H8TMI3 A0A2S2Q4T6
Pubmed
19121390
26354079
22118469
20075255
26999592
20798317
+ More
26483478 21347285 24508170 30249741 21719571 28648823 21282665 24438588 20966253 12364791 14747013 17210077 25244985 17994087 17550304 20920257 23761445 11040216 10731132 12537572 12537569 11040217 12490954 12782658 12934007 15380072 15056674 14730313 15741180 17372656 18327897 24495485 22936249 15632085 25348373 26108605 18362917 19820115 25830018 25315136 23537049 20566863 25474469 26334808 27129103
26483478 21347285 24508170 30249741 21719571 28648823 21282665 24438588 20966253 12364791 14747013 17210077 25244985 17994087 17550304 20920257 23761445 11040216 10731132 12537572 12537569 11040217 12490954 12782658 12934007 15380072 15056674 14730313 15741180 17372656 18327897 24495485 22936249 15632085 25348373 26108605 18362917 19820115 25830018 25315136 23537049 20566863 25474469 26334808 27129103
EMBL
BABH01034508
ODYU01000710
SOQ35934.1
KQ461191
KPJ07079.1
KQ458860
+ More
KPJ04782.1 AGBW02014991 OWR40770.1 NWSH01000225 PCG78232.1 AAZX01006342 GDUN01000827 JAN95092.1 GL445701 EFN89194.1 JXUM01138099 JXUM01138100 JXUM01141537 KQ569265 KXJ68689.1 KQ977876 KYM99100.1 KQ434839 KZC07992.1 GL437059 EFN71210.1 KQ435721 KOX78553.1 ADTU01000840 KK107337 QOIP01000002 EZA52451.1 RLU25886.1 GL888033 EGI69079.1 NNAY01003830 OXU18791.1 GL764736 EFZ17213.1 KQ983012 KYQ48347.1 KQ976423 KYM88781.1 AJWK01034746 ATLV01023054 KE525340 KFB48361.1 UFQT01002082 SSX32601.1 GANO01001663 JAB58208.1 AAAB01008948 EAU76692.2 APCN01004686 AXCM01002330 AXCM01002331 CM000158 EDW91497.1 NNAY01003450 OXU19398.1 KQ414666 KOC65090.1 AXCN02000986 CH954179 EDV55209.1 GFDL01013848 JAV21197.1 CH963850 EDW74795.1 CH940648 EDW61138.1 CH933808 EDW09832.1 DS231851 EDS37271.1 GGFM01001445 MBW22196.1 ADMH02002156 ETN58233.1 CP012524 ALC42603.1 AF222864 AE013599 AY069140 BT001668 BT023840 GAMC01002697 JAC03859.1 CH480816 EDW48601.1 CM002911 KMY95116.1 GGFJ01001179 MBW50320.1 CVRI01000047 CRK97601.1 CM000071 EAL24790.1 CH479181 EDW31501.1 OUUW01000001 SPP73143.1 CH902619 EDV35705.1 GGFJ01001180 MBW50321.1 GGFK01003701 MBW37022.1 CH916367 EDW01884.1 GAKP01005039 JAC53913.1 JRES01000153 KNC33767.1 AJVK01025008 AJVK01025009 AJVK01025010 KQ971345 EFA05446.1 CCAG010020342 JXJN01002477 GBYB01002803 JAG72570.1 GBXI01014489 JAC99802.1 NEVH01019959 PNF22323.1 KB632429 ERL95479.1 DS235745 EEB16219.1 KZ288318 PBC28287.1 GBYB01002804 JAG72571.1 GBBI01002728 JAC15984.1 GECL01003176 JAP02948.1 GBGD01000258 JAC88631.1 GDKW01001617 JAI54978.1 GFTR01008536 JAW07890.1 ACPB03000761 GFXV01003336 MBW15141.1 GGMS01003536 MBY72739.1
KPJ04782.1 AGBW02014991 OWR40770.1 NWSH01000225 PCG78232.1 AAZX01006342 GDUN01000827 JAN95092.1 GL445701 EFN89194.1 JXUM01138099 JXUM01138100 JXUM01141537 KQ569265 KXJ68689.1 KQ977876 KYM99100.1 KQ434839 KZC07992.1 GL437059 EFN71210.1 KQ435721 KOX78553.1 ADTU01000840 KK107337 QOIP01000002 EZA52451.1 RLU25886.1 GL888033 EGI69079.1 NNAY01003830 OXU18791.1 GL764736 EFZ17213.1 KQ983012 KYQ48347.1 KQ976423 KYM88781.1 AJWK01034746 ATLV01023054 KE525340 KFB48361.1 UFQT01002082 SSX32601.1 GANO01001663 JAB58208.1 AAAB01008948 EAU76692.2 APCN01004686 AXCM01002330 AXCM01002331 CM000158 EDW91497.1 NNAY01003450 OXU19398.1 KQ414666 KOC65090.1 AXCN02000986 CH954179 EDV55209.1 GFDL01013848 JAV21197.1 CH963850 EDW74795.1 CH940648 EDW61138.1 CH933808 EDW09832.1 DS231851 EDS37271.1 GGFM01001445 MBW22196.1 ADMH02002156 ETN58233.1 CP012524 ALC42603.1 AF222864 AE013599 AY069140 BT001668 BT023840 GAMC01002697 JAC03859.1 CH480816 EDW48601.1 CM002911 KMY95116.1 GGFJ01001179 MBW50320.1 CVRI01000047 CRK97601.1 CM000071 EAL24790.1 CH479181 EDW31501.1 OUUW01000001 SPP73143.1 CH902619 EDV35705.1 GGFJ01001180 MBW50321.1 GGFK01003701 MBW37022.1 CH916367 EDW01884.1 GAKP01005039 JAC53913.1 JRES01000153 KNC33767.1 AJVK01025008 AJVK01025009 AJVK01025010 KQ971345 EFA05446.1 CCAG010020342 JXJN01002477 GBYB01002803 JAG72570.1 GBXI01014489 JAC99802.1 NEVH01019959 PNF22323.1 KB632429 ERL95479.1 DS235745 EEB16219.1 KZ288318 PBC28287.1 GBYB01002804 JAG72571.1 GBBI01002728 JAC15984.1 GECL01003176 JAP02948.1 GBGD01000258 JAC88631.1 GDKW01001617 JAI54978.1 GFTR01008536 JAW07890.1 ACPB03000761 GFXV01003336 MBW15141.1 GGMS01003536 MBY72739.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000002358
+ More
UP000008237 UP000069940 UP000249989 UP000095300 UP000078542 UP000076502 UP000000311 UP000053105 UP000005205 UP000053097 UP000279307 UP000007755 UP000215335 UP000075809 UP000078540 UP000092461 UP000075885 UP000030765 UP000075881 UP000075902 UP000075882 UP000007062 UP000075840 UP000076407 UP000075903 UP000075900 UP000075883 UP000076408 UP000075884 UP000002282 UP000053825 UP000075886 UP000075880 UP000008711 UP000007798 UP000075920 UP000008792 UP000009192 UP000002320 UP000069272 UP000000673 UP000092553 UP000000803 UP000001292 UP000192221 UP000183832 UP000001819 UP000008744 UP000268350 UP000007801 UP000001070 UP000037069 UP000092462 UP000007266 UP000091820 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000095301 UP000235965 UP000030742 UP000005203 UP000009046 UP000242457 UP000015103
UP000008237 UP000069940 UP000249989 UP000095300 UP000078542 UP000076502 UP000000311 UP000053105 UP000005205 UP000053097 UP000279307 UP000007755 UP000215335 UP000075809 UP000078540 UP000092461 UP000075885 UP000030765 UP000075881 UP000075902 UP000075882 UP000007062 UP000075840 UP000076407 UP000075903 UP000075900 UP000075883 UP000076408 UP000075884 UP000002282 UP000053825 UP000075886 UP000075880 UP000008711 UP000007798 UP000075920 UP000008792 UP000009192 UP000002320 UP000069272 UP000000673 UP000092553 UP000000803 UP000001292 UP000192221 UP000183832 UP000001819 UP000008744 UP000268350 UP000007801 UP000001070 UP000037069 UP000092462 UP000007266 UP000091820 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000095301 UP000235965 UP000030742 UP000005203 UP000009046 UP000242457 UP000015103
Interpro
IPR005824
KOW
+ More
IPR039385 NGN_Euk
IPR024945 Spt5_C_dom
IPR036735 NGN_dom_sf
IPR041976 KOW_Spt5_3
IPR008991 Translation_prot_SH3-like_sf
IPR039659 SPT5
IPR006645 NGN_dom
IPR041973 KOW_Spt5_1
IPR041975 KOW_Spt5_2
IPR041978 KOW_Spt5_5
IPR022581 Spt5_N
IPR017071 TF_Spt5_eukaryote
IPR041980 KOW_Spt5_6
IPR041977 KOW_Spt5_4
IPR014722 Rib_L2_dom2
IPR005100 NGN-domain
IPR000684 RNA_pol_II_repeat_euk
IPR036748 MTH938-like_sf
IPR021867 Bmt2/SAMTOR
IPR007523 NDUFAF3/AAMDC
IPR029063 SAM-dependent_MTases
IPR034095 NDUF3
IPR001781 Znf_LIM
IPR002931 Transglutaminase-like
IPR038765 Papain-like_cys_pep_sf
IPR039385 NGN_Euk
IPR024945 Spt5_C_dom
IPR036735 NGN_dom_sf
IPR041976 KOW_Spt5_3
IPR008991 Translation_prot_SH3-like_sf
IPR039659 SPT5
IPR006645 NGN_dom
IPR041973 KOW_Spt5_1
IPR041975 KOW_Spt5_2
IPR041978 KOW_Spt5_5
IPR022581 Spt5_N
IPR017071 TF_Spt5_eukaryote
IPR041980 KOW_Spt5_6
IPR041977 KOW_Spt5_4
IPR014722 Rib_L2_dom2
IPR005100 NGN-domain
IPR000684 RNA_pol_II_repeat_euk
IPR036748 MTH938-like_sf
IPR021867 Bmt2/SAMTOR
IPR007523 NDUFAF3/AAMDC
IPR029063 SAM-dependent_MTases
IPR034095 NDUF3
IPR001781 Znf_LIM
IPR002931 Transglutaminase-like
IPR038765 Papain-like_cys_pep_sf
Gene 3D
CDD
ProteinModelPortal
H9J2M2
A0A2H1V6P2
A0A194QP01
A0A194QGZ7
A0A212EH19
A0A2A4K3K3
+ More
K7J5S6 A0A1S4FZD5 A0A0P6IVB5 E2B534 A0A182G1W4 A0A182G3H6 A0A1I8NZL8 A0A195CEG6 A0A154P7Y4 E2A5T8 A0A0M9A996 A0A158NYZ9 A0A026W9W4 F4W9U3 A0A232EKF0 E9IQE3 A0A151WKL0 A0A195BQD0 A0A1B0EUS0 A0A182PF58 A0A084WDR7 A0A336MSR5 A0A182K2J7 U5EVY9 A0A182U6J2 A0A182KYR8 A0NE83 A0A182HQS7 A0A182XJE1 A0A182VIZ1 A0A182RUU0 A0A182M7H4 A0A182YLF7 A0A182N1S7 B4PB84 A0A232EM50 A0A0L7R2I7 A0A182QPP4 A0A182JHT9 A0A1S4J862 B3NJV0 A0A1Q3F0W5 B4MRQ6 A0A182WM74 B4LNW9 B4KME4 B0W6Z7 A0A2M3Z117 A0A182FLB7 W5J4B3 A0A0M3QVL5 Q9V460 W8C3N1 B4HPG3 A0A0J9RGS1 A0A2M4BBB8 A0A1W4VGR7 A0A1J1IGM6 Q292S1 B4GCQ3 A0A3B0J2C6 B3MGG2 A0A2M4BBF7 A0A2M4A8G2 B4J626 A0A034WJE4 A0A0L0CNG6 A0A1B0D500 D2A605 A0A1A9WAC5 A0A1B0FL32 A0A1A9YJP3 A0A1A9UYD5 A0A1B0ARK9 A0A0C9PNW0 A0A0A1WN47 A0A1A9ZRD1 A0A1I8M549 A0A2J7Q156 U4UN14 A0A088A942 E0VS63 A0A2A3E981 A0A0C9QH50 A0A023F4S8 A0A0V0G4H6 A0A069DXS5 A0A0P4VJ54 A0A224XHF9 T1I785 A0A2H8TMI3 A0A2S2Q4T6
K7J5S6 A0A1S4FZD5 A0A0P6IVB5 E2B534 A0A182G1W4 A0A182G3H6 A0A1I8NZL8 A0A195CEG6 A0A154P7Y4 E2A5T8 A0A0M9A996 A0A158NYZ9 A0A026W9W4 F4W9U3 A0A232EKF0 E9IQE3 A0A151WKL0 A0A195BQD0 A0A1B0EUS0 A0A182PF58 A0A084WDR7 A0A336MSR5 A0A182K2J7 U5EVY9 A0A182U6J2 A0A182KYR8 A0NE83 A0A182HQS7 A0A182XJE1 A0A182VIZ1 A0A182RUU0 A0A182M7H4 A0A182YLF7 A0A182N1S7 B4PB84 A0A232EM50 A0A0L7R2I7 A0A182QPP4 A0A182JHT9 A0A1S4J862 B3NJV0 A0A1Q3F0W5 B4MRQ6 A0A182WM74 B4LNW9 B4KME4 B0W6Z7 A0A2M3Z117 A0A182FLB7 W5J4B3 A0A0M3QVL5 Q9V460 W8C3N1 B4HPG3 A0A0J9RGS1 A0A2M4BBB8 A0A1W4VGR7 A0A1J1IGM6 Q292S1 B4GCQ3 A0A3B0J2C6 B3MGG2 A0A2M4BBF7 A0A2M4A8G2 B4J626 A0A034WJE4 A0A0L0CNG6 A0A1B0D500 D2A605 A0A1A9WAC5 A0A1B0FL32 A0A1A9YJP3 A0A1A9UYD5 A0A1B0ARK9 A0A0C9PNW0 A0A0A1WN47 A0A1A9ZRD1 A0A1I8M549 A0A2J7Q156 U4UN14 A0A088A942 E0VS63 A0A2A3E981 A0A0C9QH50 A0A023F4S8 A0A0V0G4H6 A0A069DXS5 A0A0P4VJ54 A0A224XHF9 T1I785 A0A2H8TMI3 A0A2S2Q4T6
PDB
6GML
E-value=0,
Score=2138
Ontologies
GO
GO:0006357
GO:0032784
GO:0005634
GO:0003746
GO:0003677
GO:0006366
GO:0005665
GO:0008168
GO:0032981
GO:0006368
GO:0003729
GO:0032044
GO:0003682
GO:0000993
GO:0045944
GO:0032785
GO:0044877
GO:0005705
GO:0000122
GO:0005703
GO:0007549
GO:0008023
GO:0032786
GO:0046982
GO:0005700
GO:0006355
GO:0005515
GO:0008017
GO:0006289
GO:0004518
GO:0006281
GO:0048384
PANTHER
Topology
Subcellular location
Nucleus
Chromosome
Chromosome
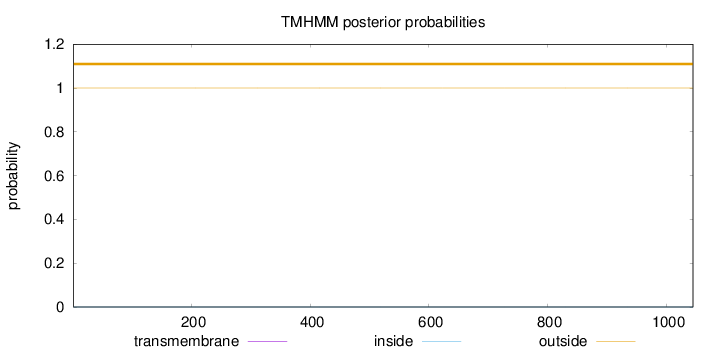
Length:
1045
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00125
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 1045
Population Genetic Test Statistics
Pi
2.314263
Theta
5.021368
Tajima's D
-1.663896
CLR
340.015107
CSRT
0.0343482825858707
Interpretation
Possibly Positive selection
