Gene
KWMTBOMO02325 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003788
Annotation
PREDICTED:_long-chain-fatty-acid--CoA_ligase_4_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.459 Mitochondrial Reliability : 1.876
Sequence
CDS
ATGTTCATAGACGAAAACGAAACGCCCGACTCATGGTGGGTGTCGGCGGTGCTAAGGACCATACGGACCGTCGCGTTTGTCTTCGACGTCATCACGTTCCCGATCCACCTCCTCGTCCAGAGACCTTGGAGGAAACGTGCCTTGTCAAGAAGAATAAAGGCTCGTATAATAAGCAGTACCGAGAACAGCATAACGATTAGTCCGCTAACGCCACCCTGCGACCTACACCTGCGTCTGGTCCGCGATGGCGTCACCACCATGGAGGGCATGCTCCGCGCCGCCACCCAGCGCTGGGGGCAGCGGCCCTGTCTGGGGACGCGCGCCGTGCTCAGCGAGGAGGACGAACCGCAACCAAACGGACGAGTCTTTAAGAAGTTCAAAATGGGCGAGTACATATGGCGGACGTACGCGGACGTAGAGCGTGAGGCGCGAGAGTTCGGCGCGGGGCTGCGCGCGGTGGGCTGCGAACCGCGGCGCAACGTCGTCATTTTCGCAGAGACGCGCGCCGAGTGGATGCTCGCCGCGCACGGCTGCTTCACGCAGAGCATCCCCGTTGTCACAATCTATGCCACACTGGGAGACGAAGCTATCGCTCACGGCATCAACGAGACCGAAGTGTCCACGGTCATCACCACGCACGATCTCCTGCCCAAGTTCAAGAAGATTCTCGCGAAGACGCCCAAAGTGGAGACCATCATCTACATGGAGGACCAGCTCAAGCCGACCGACACGCAGGGCTTCAAGGAGGGAATCAAGATCGTGAGCTACAAGAATGTCGTGGAACTCGGCAGGGAGTCTAAGATCGAGAGCGTGCGGCCCGGCGCGGCGGACACGGCCATCATCATGTACACGTCGGGCTCCACGGGCGTGCCCAAGGGCGTGCTGCTGTCGCACCGCAACATGCTGGCCACGCTCAAGGCCTTCGCCGACGCCATCCCCATCTACGAGGGCGACGTGCTCATGGGCTTCCTGCCGCTGGCGCACGTGTTCGAGCTGCTCGCCGAGAACCTCTGCATCATCGGAGGCGTGCCCATCGGCTACTCCACTCCGCTGACGATGCTGGACTCGTCCAGCAAGATCATGAAGGGGACGAAGGGCGACGCCACCACGCTGAGGCCGACCTGCATCACCACCGTGCCGTTGATAATGGACCGCATCAGCAAGGGCATCACGGACAAGGTGAGTCGCAGCGGGGAGCTGGCCGCCGCCGTGTTCAAGTGGAGCTACAGCTACAAGCAGACCTGGATGCGGCGCGGCTACGACACGCCCGTGCTGGACTACCTGATATTCCGCAAAGTGGGCGGGCTGCTGGGCGGCCGCGTGCGCCTCATGATCTCGGGCGGCGCGCCCCTGTCGCCTGACACACACCTACAGGTGAAGATCTGCCTGTGCTGCTCCGTGGTGGCGGGCTACGGGCTCACGGAGACCACGTCGTGCGGCACCGTCATGGACCCGCACGACCGCTCCACGGGCCGCGTGGGCGCCCCGGCCGCCGGCGCGCTGCTGGCGCTGCGGGACTGGGACGAGGGCAACTACCGCGTCACCAACAAGCCGCACCCGCAGGGCGAGATCATAATCGGCGGTGACTGCGTCGCCGAGGGGTACTACAAGAACCCGCAGAAGACGAAGGAGGAATTCATCGACGCCGAGGGGAGACGCTGGTTCAAATCCGGAGATATCGGGGAGCTGCATGACGATGGATGTATTAAGATTATTGACCGCAAGAAGGACCTGGTGAAGCTGCAGGCCGGCGAGTACGTGTCGCTGGGCAAGGTGGAGGCCGAGCTGAAGACGTGCCCCATCGTGGAGAACATCTGCGTGTACGGCGACAGCAGCAAGTCGCACACCGTGGCGCTGGTCGTGCCCAACGCGCCGCGCCTGGCCGCGCTCGCCCAGCGCCTGCAGCTGCCCGCGGGCGCCGACCCCCCCGCGCTGTACGCGCAGCCCGCGCTGGAGCGCGCCGTGCTGGCCGAGCTGGCCGAGCACGCGCGCAAGTGCGGCCTCGAGAAGTTCGAGGTGCCCGCCGCCGTCAAGCTGTGCACCGAGGTCTGGTCGCCCGACATGGGGCTGGTCACCGCCGCCTTCAAGATCAAGCGCAAGGACATCCAGGAGCGGTACAAGGAGGACATCAAGCGGATGTACGCGTCCTGA
Protein
MFIDENETPDSWWVSAVLRTIRTVAFVFDVITFPIHLLVQRPWRKRALSRRIKARIISSTENSITISPLTPPCDLHLRLVRDGVTTMEGMLRAATQRWGQRPCLGTRAVLSEEDEPQPNGRVFKKFKMGEYIWRTYADVEREAREFGAGLRAVGCEPRRNVVIFAETRAEWMLAAHGCFTQSIPVVTIYATLGDEAIAHGINETEVSTVITTHDLLPKFKKILAKTPKVETIIYMEDQLKPTDTQGFKEGIKIVSYKNVVELGRESKIESVRPGAADTAIIMYTSGSTGVPKGVLLSHRNMLATLKAFADAIPIYEGDVLMGFLPLAHVFELLAENLCIIGGVPIGYSTPLTMLDSSSKIMKGTKGDATTLRPTCITTVPLIMDRISKGITDKVSRSGELAAAVFKWSYSYKQTWMRRGYDTPVLDYLIFRKVGGLLGGRVRLMISGGAPLSPDTHLQVKICLCCSVVAGYGLTETTSCGTVMDPHDRSTGRVGAPAAGALLALRDWDEGNYRVTNKPHPQGEIIIGGDCVAEGYYKNPQKTKEEFIDAEGRRWFKSGDIGELHDDGCIKIIDRKKDLVKLQAGEYVSLGKVEAELKTCPIVENICVYGDSSKSHTVALVVPNAPRLAALAQRLQLPAGADPPALYAQPALERAVLAELAEHARKCGLEKFEVPAAVKLCTEVWSPDMGLVTAAFKIKRKDIQERYKEDIKRMYAS
Summary
Uniprot
H9J2Q1
A0A194QQ38
A0A2A4K1U3
A0A1A9Z1V8
E2AYR1
A0A1A9V1L5
+ More
A0A069DWY9 A0A1A9YD05 A0A1B0C0T5 A0A1Y1MN63 A0A224XJI1 T1PFX0 A0A1I8NID3 A0A158NCE8 D3TR21 A0A151WMR0 A0A1B6E8J8 E2BBW6 A0A0L0BR37 A0A3L8DHP9 Q28Y67 A0A1Y1MFT9 A0A151JQZ1 A0A1B0F9R3 A0A195FAH7 A0A151ICB8 A0A026X1X7 T1I0F4 A0A034W989 A0A0K8U4E7 A0A1L8DZ15 A0A2J7QBQ0 A0A2J7QBN9 A0A0P4W361 A0A3B0J0R5 A0A1A9W3R7 W8B3H8 A0A0J7L6J6 B4MPM8 A0A0Q9WG92 A0A067QQW2 A0A0M4EAA3 A0A0K8TNA5 A0A034W7L0 A0A195BY92 B4GGL7 A0A0A1XED0 B4KMY4 A0A0R3NUJ0 A0A1L8DZ12 A0A3B0JT50 E9INK5 W8BQ32 A0A1L8DYX3 A0A0Q9W451 A0A182WVN2 A0A2C9GMU5 F5HLC1 A0A0L7RG48 E0VY58 A0A1W4WKU7 F4WWY9 A0A182UUS7 A0A0Q9XJC4 N6WCC2 A0A0A1XHW8 A0A0A1XNL1 B4LNB5 A0A1Y1MI90 A0A1Q3FY57 A0A2C9H6R6 A0A1Q3FY73 F5HLC2 A0A182I9B2 A0A182UG18 A0A2C9H6R4 A0A3B0JLD2 A0A2C9GN04 A0A182KXP0 Q7QAM6 A0A154PPC9 A0A182KEN9 A0A0N0U4J8 A0A2M4BFK7 A0A0R1DRJ7 A0A182MGC3 A0A182QJK3 A0A2M4A346 A0A1Q3G3M4 A0A2M3Z0L9 A0A182REW1 A0A182P901 A0A0Q5WKV4 Q8T3L1 A0A1S4FRU9 B4QG64 W5JAD8 A0A023EXN4 A0A139WH95 A0A084VW14
A0A069DWY9 A0A1A9YD05 A0A1B0C0T5 A0A1Y1MN63 A0A224XJI1 T1PFX0 A0A1I8NID3 A0A158NCE8 D3TR21 A0A151WMR0 A0A1B6E8J8 E2BBW6 A0A0L0BR37 A0A3L8DHP9 Q28Y67 A0A1Y1MFT9 A0A151JQZ1 A0A1B0F9R3 A0A195FAH7 A0A151ICB8 A0A026X1X7 T1I0F4 A0A034W989 A0A0K8U4E7 A0A1L8DZ15 A0A2J7QBQ0 A0A2J7QBN9 A0A0P4W361 A0A3B0J0R5 A0A1A9W3R7 W8B3H8 A0A0J7L6J6 B4MPM8 A0A0Q9WG92 A0A067QQW2 A0A0M4EAA3 A0A0K8TNA5 A0A034W7L0 A0A195BY92 B4GGL7 A0A0A1XED0 B4KMY4 A0A0R3NUJ0 A0A1L8DZ12 A0A3B0JT50 E9INK5 W8BQ32 A0A1L8DYX3 A0A0Q9W451 A0A182WVN2 A0A2C9GMU5 F5HLC1 A0A0L7RG48 E0VY58 A0A1W4WKU7 F4WWY9 A0A182UUS7 A0A0Q9XJC4 N6WCC2 A0A0A1XHW8 A0A0A1XNL1 B4LNB5 A0A1Y1MI90 A0A1Q3FY57 A0A2C9H6R6 A0A1Q3FY73 F5HLC2 A0A182I9B2 A0A182UG18 A0A2C9H6R4 A0A3B0JLD2 A0A2C9GN04 A0A182KXP0 Q7QAM6 A0A154PPC9 A0A182KEN9 A0A0N0U4J8 A0A2M4BFK7 A0A0R1DRJ7 A0A182MGC3 A0A182QJK3 A0A2M4A346 A0A1Q3G3M4 A0A2M3Z0L9 A0A182REW1 A0A182P901 A0A0Q5WKV4 Q8T3L1 A0A1S4FRU9 B4QG64 W5JAD8 A0A023EXN4 A0A139WH95 A0A084VW14
Pubmed
19121390
26354079
20798317
26334808
28004739
25315136
+ More
21347285 20353571 26108605 30249741 15632085 17994087 24508170 25348373 27129103 24495485 18057021 24845553 26369729 25830018 23185243 21282665 12364791 14747013 17210077 20566863 21719571 20966253 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20920257 23761445 24945155 18362917 19820115 24438588
21347285 20353571 26108605 30249741 15632085 17994087 24508170 25348373 27129103 24495485 18057021 24845553 26369729 25830018 23185243 21282665 12364791 14747013 17210077 20566863 21719571 20966253 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20920257 23761445 24945155 18362917 19820115 24438588
EMBL
BABH01034503
BABH01034504
BABH01034505
BABH01034506
BABH01034507
KQ461191
+ More
KPJ07080.1 NWSH01000225 PCG78235.1 GL443983 EFN61424.1 GBGD01000667 JAC88222.1 JXJN01023749 JXJN01023750 GEZM01032304 JAV84677.1 GFTR01007806 JAW08620.1 KA647569 AFP62198.1 ADTU01011838 EZ423873 ADD20149.1 KQ982934 KYQ49159.1 GEDC01029969 GEDC01003038 JAS07329.1 JAS34260.1 GL447175 EFN86790.1 JRES01001589 KNC21699.1 QOIP01000008 RLU19683.1 CM000071 EAL26098.3 GEZM01032302 JAV84679.1 KQ978619 KYN29777.1 CCAG010011069 CCAG010011070 KQ981727 KYN37049.1 KQ978076 KYM97216.1 KK107034 EZA62068.1 ACPB03006604 KT328602 ALT07199.1 GAKP01008282 GAKP01008281 GAKP01008277 GAKP01008272 GAKP01008270 GAKP01008268 JAC50680.1 GDHF01033727 GDHF01030886 GDHF01021668 JAI18587.1 JAI21428.1 JAI30646.1 GFDF01002400 JAV11684.1 NEVH01016296 PNF25998.1 PNF25996.1 GDKW01000673 JAI55922.1 OUUW01000001 SPP74307.1 GAMC01014952 GAMC01014949 JAB91603.1 LBMM01000501 KMQ98198.1 CH963849 EDW74067.1 CH940648 KRF79749.1 KRF79750.1 KK853129 KDR10971.1 CP012524 ALC41958.1 GDAI01001749 JAI15854.1 GAKP01008274 GAKP01008273 GAKP01008269 JAC50679.1 KQ976396 KYM92906.1 CH479183 EDW35637.1 GBXI01005454 GBXI01005112 JAD08838.1 JAD09180.1 CH933808 EDW09906.1 KRT02844.1 KRT02845.1 GFDF01002427 JAV11657.1 SPP74308.1 GL764306 EFZ17862.1 GAMC01014951 JAB91604.1 GFDF01002426 JAV11658.1 KRF79751.1 KRF79752.1 APCN01003249 APCN01003250 AAAB01008888 EGK97083.1 KQ414598 KOC69808.1 DS235843 EEB18314.1 GL888417 EGI61232.1 KRG04974.1 KRG04975.1 ENO01794.2 KRT02842.1 KRT02843.1 GBXI01003782 JAD10510.1 GBXI01001696 JAD12596.1 EDW61067.1 GEZM01032303 JAV84678.1 GFDL01002558 JAV32487.1 GFDL01002559 JAV32486.1 EGK97082.1 SPP74309.1 EAA08767.2 KQ435007 KZC13597.1 KQ435830 KOX71758.1 GGFJ01002683 MBW51824.1 CM000157 KRJ98370.1 KRJ98379.1 AXCM01007567 AXCN02000282 GGFK01001912 MBW35233.1 GFDL01000654 JAV34391.1 GGFM01001326 MBW22077.1 CH954177 KQS70862.1 AE013599 AY094958 AAF59061.2 AAG22300.2 AAM11311.1 AAX52718.1 AAX52719.1 CM000362 CM002911 EDX06214.1 KMY92296.1 ADMH02002036 ETN59825.1 GAPW01000354 JAC13244.1 KQ971343 KYB27254.1 ATLV01017372 KE525164 KFB42158.1
KPJ07080.1 NWSH01000225 PCG78235.1 GL443983 EFN61424.1 GBGD01000667 JAC88222.1 JXJN01023749 JXJN01023750 GEZM01032304 JAV84677.1 GFTR01007806 JAW08620.1 KA647569 AFP62198.1 ADTU01011838 EZ423873 ADD20149.1 KQ982934 KYQ49159.1 GEDC01029969 GEDC01003038 JAS07329.1 JAS34260.1 GL447175 EFN86790.1 JRES01001589 KNC21699.1 QOIP01000008 RLU19683.1 CM000071 EAL26098.3 GEZM01032302 JAV84679.1 KQ978619 KYN29777.1 CCAG010011069 CCAG010011070 KQ981727 KYN37049.1 KQ978076 KYM97216.1 KK107034 EZA62068.1 ACPB03006604 KT328602 ALT07199.1 GAKP01008282 GAKP01008281 GAKP01008277 GAKP01008272 GAKP01008270 GAKP01008268 JAC50680.1 GDHF01033727 GDHF01030886 GDHF01021668 JAI18587.1 JAI21428.1 JAI30646.1 GFDF01002400 JAV11684.1 NEVH01016296 PNF25998.1 PNF25996.1 GDKW01000673 JAI55922.1 OUUW01000001 SPP74307.1 GAMC01014952 GAMC01014949 JAB91603.1 LBMM01000501 KMQ98198.1 CH963849 EDW74067.1 CH940648 KRF79749.1 KRF79750.1 KK853129 KDR10971.1 CP012524 ALC41958.1 GDAI01001749 JAI15854.1 GAKP01008274 GAKP01008273 GAKP01008269 JAC50679.1 KQ976396 KYM92906.1 CH479183 EDW35637.1 GBXI01005454 GBXI01005112 JAD08838.1 JAD09180.1 CH933808 EDW09906.1 KRT02844.1 KRT02845.1 GFDF01002427 JAV11657.1 SPP74308.1 GL764306 EFZ17862.1 GAMC01014951 JAB91604.1 GFDF01002426 JAV11658.1 KRF79751.1 KRF79752.1 APCN01003249 APCN01003250 AAAB01008888 EGK97083.1 KQ414598 KOC69808.1 DS235843 EEB18314.1 GL888417 EGI61232.1 KRG04974.1 KRG04975.1 ENO01794.2 KRT02842.1 KRT02843.1 GBXI01003782 JAD10510.1 GBXI01001696 JAD12596.1 EDW61067.1 GEZM01032303 JAV84678.1 GFDL01002558 JAV32487.1 GFDL01002559 JAV32486.1 EGK97082.1 SPP74309.1 EAA08767.2 KQ435007 KZC13597.1 KQ435830 KOX71758.1 GGFJ01002683 MBW51824.1 CM000157 KRJ98370.1 KRJ98379.1 AXCM01007567 AXCN02000282 GGFK01001912 MBW35233.1 GFDL01000654 JAV34391.1 GGFM01001326 MBW22077.1 CH954177 KQS70862.1 AE013599 AY094958 AAF59061.2 AAG22300.2 AAM11311.1 AAX52718.1 AAX52719.1 CM000362 CM002911 EDX06214.1 KMY92296.1 ADMH02002036 ETN59825.1 GAPW01000354 JAC13244.1 KQ971343 KYB27254.1 ATLV01017372 KE525164 KFB42158.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000092445
UP000000311
UP000078200
+ More
UP000092443 UP000092460 UP000095301 UP000005205 UP000075809 UP000008237 UP000037069 UP000279307 UP000001819 UP000078492 UP000092444 UP000078541 UP000078542 UP000053097 UP000015103 UP000235965 UP000268350 UP000091820 UP000036403 UP000007798 UP000008792 UP000027135 UP000092553 UP000078540 UP000008744 UP000009192 UP000076407 UP000075840 UP000007062 UP000053825 UP000009046 UP000192223 UP000007755 UP000075903 UP000075902 UP000075882 UP000076502 UP000075881 UP000053105 UP000002282 UP000075883 UP000075886 UP000075900 UP000075885 UP000008711 UP000000803 UP000000304 UP000000673 UP000007266 UP000030765
UP000092443 UP000092460 UP000095301 UP000005205 UP000075809 UP000008237 UP000037069 UP000279307 UP000001819 UP000078492 UP000092444 UP000078541 UP000078542 UP000053097 UP000015103 UP000235965 UP000268350 UP000091820 UP000036403 UP000007798 UP000008792 UP000027135 UP000092553 UP000078540 UP000008744 UP000009192 UP000076407 UP000075840 UP000007062 UP000053825 UP000009046 UP000192223 UP000007755 UP000075903 UP000075902 UP000075882 UP000076502 UP000075881 UP000053105 UP000002282 UP000075883 UP000075886 UP000075900 UP000075885 UP000008711 UP000000803 UP000000304 UP000000673 UP000007266 UP000030765
Interpro
Gene 3D
ProteinModelPortal
H9J2Q1
A0A194QQ38
A0A2A4K1U3
A0A1A9Z1V8
E2AYR1
A0A1A9V1L5
+ More
A0A069DWY9 A0A1A9YD05 A0A1B0C0T5 A0A1Y1MN63 A0A224XJI1 T1PFX0 A0A1I8NID3 A0A158NCE8 D3TR21 A0A151WMR0 A0A1B6E8J8 E2BBW6 A0A0L0BR37 A0A3L8DHP9 Q28Y67 A0A1Y1MFT9 A0A151JQZ1 A0A1B0F9R3 A0A195FAH7 A0A151ICB8 A0A026X1X7 T1I0F4 A0A034W989 A0A0K8U4E7 A0A1L8DZ15 A0A2J7QBQ0 A0A2J7QBN9 A0A0P4W361 A0A3B0J0R5 A0A1A9W3R7 W8B3H8 A0A0J7L6J6 B4MPM8 A0A0Q9WG92 A0A067QQW2 A0A0M4EAA3 A0A0K8TNA5 A0A034W7L0 A0A195BY92 B4GGL7 A0A0A1XED0 B4KMY4 A0A0R3NUJ0 A0A1L8DZ12 A0A3B0JT50 E9INK5 W8BQ32 A0A1L8DYX3 A0A0Q9W451 A0A182WVN2 A0A2C9GMU5 F5HLC1 A0A0L7RG48 E0VY58 A0A1W4WKU7 F4WWY9 A0A182UUS7 A0A0Q9XJC4 N6WCC2 A0A0A1XHW8 A0A0A1XNL1 B4LNB5 A0A1Y1MI90 A0A1Q3FY57 A0A2C9H6R6 A0A1Q3FY73 F5HLC2 A0A182I9B2 A0A182UG18 A0A2C9H6R4 A0A3B0JLD2 A0A2C9GN04 A0A182KXP0 Q7QAM6 A0A154PPC9 A0A182KEN9 A0A0N0U4J8 A0A2M4BFK7 A0A0R1DRJ7 A0A182MGC3 A0A182QJK3 A0A2M4A346 A0A1Q3G3M4 A0A2M3Z0L9 A0A182REW1 A0A182P901 A0A0Q5WKV4 Q8T3L1 A0A1S4FRU9 B4QG64 W5JAD8 A0A023EXN4 A0A139WH95 A0A084VW14
A0A069DWY9 A0A1A9YD05 A0A1B0C0T5 A0A1Y1MN63 A0A224XJI1 T1PFX0 A0A1I8NID3 A0A158NCE8 D3TR21 A0A151WMR0 A0A1B6E8J8 E2BBW6 A0A0L0BR37 A0A3L8DHP9 Q28Y67 A0A1Y1MFT9 A0A151JQZ1 A0A1B0F9R3 A0A195FAH7 A0A151ICB8 A0A026X1X7 T1I0F4 A0A034W989 A0A0K8U4E7 A0A1L8DZ15 A0A2J7QBQ0 A0A2J7QBN9 A0A0P4W361 A0A3B0J0R5 A0A1A9W3R7 W8B3H8 A0A0J7L6J6 B4MPM8 A0A0Q9WG92 A0A067QQW2 A0A0M4EAA3 A0A0K8TNA5 A0A034W7L0 A0A195BY92 B4GGL7 A0A0A1XED0 B4KMY4 A0A0R3NUJ0 A0A1L8DZ12 A0A3B0JT50 E9INK5 W8BQ32 A0A1L8DYX3 A0A0Q9W451 A0A182WVN2 A0A2C9GMU5 F5HLC1 A0A0L7RG48 E0VY58 A0A1W4WKU7 F4WWY9 A0A182UUS7 A0A0Q9XJC4 N6WCC2 A0A0A1XHW8 A0A0A1XNL1 B4LNB5 A0A1Y1MI90 A0A1Q3FY57 A0A2C9H6R6 A0A1Q3FY73 F5HLC2 A0A182I9B2 A0A182UG18 A0A2C9H6R4 A0A3B0JLD2 A0A2C9GN04 A0A182KXP0 Q7QAM6 A0A154PPC9 A0A182KEN9 A0A0N0U4J8 A0A2M4BFK7 A0A0R1DRJ7 A0A182MGC3 A0A182QJK3 A0A2M4A346 A0A1Q3G3M4 A0A2M3Z0L9 A0A182REW1 A0A182P901 A0A0Q5WKV4 Q8T3L1 A0A1S4FRU9 B4QG64 W5JAD8 A0A023EXN4 A0A139WH95 A0A084VW14
PDB
5MSW
E-value=3.25214e-28,
Score=314
Ontologies
PATHWAY
GO
GO:0003824
GO:0016874
GO:0102391
GO:0004467
GO:0003996
GO:0010890
GO:0005783
GO:0007411
GO:0035282
GO:0006655
GO:0005777
GO:0030424
GO:0045886
GO:0061502
GO:0030514
GO:0035338
GO:0007268
GO:0005829
GO:0090433
GO:0016021
GO:0005737
GO:0001676
GO:0007399
GO:0008152
GO:0005891
GO:0003676
GO:0008017
GO:0005634
GO:0006289
GO:0004518
GO:0006281
GO:0048384
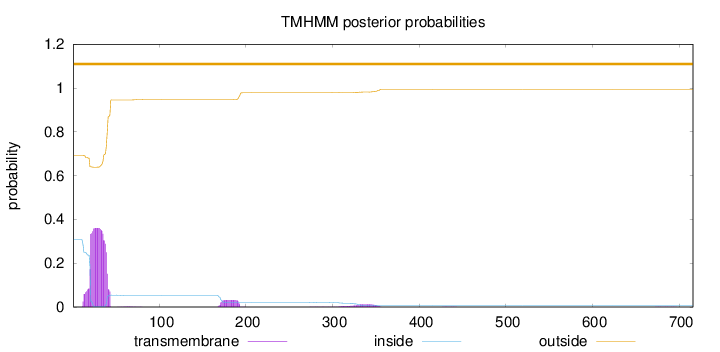
Topology
Length:
716
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.62185000000001
Exp number, first 60 AAs:
7.52447
Total prob of N-in:
0.30835
outside
1 - 716
Population Genetic Test Statistics
Pi
21.592784
Theta
17.072839
Tajima's D
-2.113263
CLR
190.560626
CSRT
0.00874956252187391
Interpretation
Possibly Positive selection
