Pre Gene Modal
BGIBMGA003762
Annotation
PREDICTED:_septum_formation_protein_Maf_isoform_X1_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 2.02
Sequence
CDS
ATGTTGCAACCAGTAATGCATGTTTTGAAAGCTAAGAAAATTGTGCTAGCCAGTGCATCACCGCGACGCGCTGAACTTATTGAAAACATCGGACTCAAAGTTAGTTTATGCCCATCATTATTTGAAGAAAACTTAGTCCCGAATGAGTTCAGCAAATTTTCAGATTTTGTTGAGGAGACAGCCCTTCACAAAGTTCTGGAGGTAGAAAAGCGTTTGACAGGAATGGGCTCTGCTCCAGACATTGTTATTGGTGCTGATACAATGGTGACACTGGATGGCGAAATGTTTGGAAAACCTACTTCAGAAGCTGAAGCATTTGTAATGCTGTCAAGATTATCAGGACGCTCACATACTGTGTACACAGGAGTGGTTGTGAAGACGGGAGAAGAGATTGTAAAATTCACTGAGTCTACTGATGTTCAGTTTGGTGAATTAGATGAACAACAAATCAGAGGGTACATTGATACGGGAGAGCCTATGGATAAAGCTGGAGGTTATGGCATCCAAGGCGTTGGAGGAACCTTTGTGGAGCGAGTTGAAGGAGATTATTTCACTGTAGTTGGCCTACCGTTATACAGATTATGCTCTGTACTCTACAAAATGCATAAACATTTAATATAA
Protein
MLQPVMHVLKAKKIVLASASPRRAELIENIGLKVSLCPSLFEENLVPNEFSKFSDFVEETALHKVLEVEKRLTGMGSAPDIVIGADTMVTLDGEMFGKPTSEAEAFVMLSRLSGRSHTVYTGVVVKTGEEIVKFTESTDVQFGELDEQQIRGYIDTGEPMDKAGGYGIQGVGGTFVERVEGDYFTVVGLPLYRLCSVLYKMHKHLI
Summary
Uniprot
H9J2M5
A0A2H1VBH3
A0A212EGZ9
A0A194QMR5
A0A194QNI4
A0A2A4J666
+ More
A0A182STQ8 A0A0L7L4X7 A0A1L8DWU9 A0A336MDU9 Q17K21 A0A1B0DB08 Q7Q8B4 B0W4W1 A0A182GQ12 A0A182U7N4 A0A182KWE8 A0A182X6A6 A0A1B6EAU3 A0A1B6EW96 A0A1B6IDR8 A0A182V2H7 A0A023ELW7 A0A1Q3F2D0 A0A2R7X2Z2 A0A182R3Z6 A0A182IBC8 A0A182VTF8 A0A0A9ZGL6 A0A0A9ZDF2 A0A182MQE3 A0A0A9ZDL2 A0A146L4B1 A0A0K8TMQ0 A0A146LE30 A0A182K804 A0A1B0G0C5 B4Q7B2 A0A034WP43 A0A1A9VA62 A0A1B0AAK6 B3MM86 A0A0R1DSI4 A0A1W4VZN9 B4HYS7 B4M9P8 A0A182R0F7 Q86BM0 A0A182NAL7 B3N7U9 A0A1B0B291 A0A2P8ZE64 A0A1A9Y1P5 A0A0K8TYW1 A0A0L0BVG0 E0VY26 A0A0A1WUM9 B4GJ33 A0A182IQ93 W8C7K5 A0A1B6JGL8 B5DK84 A0A1A9W2J9 B4JP53 A0A0M4E9S4 B4N0A0 R7UM23 A0A067RGU5 A0A3B0K919 A0A069DXV5 A0A0P4W141 R4G421 A0A2J7RNX5 A0A2M3ZJK9 A0A224XWL3 A0A1Y1K1J4 A0A1I8N6M9 A0A023F7Z4 A0A2M4BZL7 B4KEY7 A0A151X6J4 A0A2M4C068 A0A2M4CUF7 A0A2M4BZS9 A0A1B0GKL3 A0A182F3C3 A0A1L8DWM6 A0A0M9AC56 A0A2M4CU21 A0A2M4AWI7 A0A0B6Z6B4 A0A1S3H8A0 A0A1I8Q4Y8 A7SSI3 A0A0L7QW63 D6WP02 A0A2L2YKP6 A0A026WNN1 A0A3L8DV95
A0A182STQ8 A0A0L7L4X7 A0A1L8DWU9 A0A336MDU9 Q17K21 A0A1B0DB08 Q7Q8B4 B0W4W1 A0A182GQ12 A0A182U7N4 A0A182KWE8 A0A182X6A6 A0A1B6EAU3 A0A1B6EW96 A0A1B6IDR8 A0A182V2H7 A0A023ELW7 A0A1Q3F2D0 A0A2R7X2Z2 A0A182R3Z6 A0A182IBC8 A0A182VTF8 A0A0A9ZGL6 A0A0A9ZDF2 A0A182MQE3 A0A0A9ZDL2 A0A146L4B1 A0A0K8TMQ0 A0A146LE30 A0A182K804 A0A1B0G0C5 B4Q7B2 A0A034WP43 A0A1A9VA62 A0A1B0AAK6 B3MM86 A0A0R1DSI4 A0A1W4VZN9 B4HYS7 B4M9P8 A0A182R0F7 Q86BM0 A0A182NAL7 B3N7U9 A0A1B0B291 A0A2P8ZE64 A0A1A9Y1P5 A0A0K8TYW1 A0A0L0BVG0 E0VY26 A0A0A1WUM9 B4GJ33 A0A182IQ93 W8C7K5 A0A1B6JGL8 B5DK84 A0A1A9W2J9 B4JP53 A0A0M4E9S4 B4N0A0 R7UM23 A0A067RGU5 A0A3B0K919 A0A069DXV5 A0A0P4W141 R4G421 A0A2J7RNX5 A0A2M3ZJK9 A0A224XWL3 A0A1Y1K1J4 A0A1I8N6M9 A0A023F7Z4 A0A2M4BZL7 B4KEY7 A0A151X6J4 A0A2M4C068 A0A2M4CUF7 A0A2M4BZS9 A0A1B0GKL3 A0A182F3C3 A0A1L8DWM6 A0A0M9AC56 A0A2M4CU21 A0A2M4AWI7 A0A0B6Z6B4 A0A1S3H8A0 A0A1I8Q4Y8 A7SSI3 A0A0L7QW63 D6WP02 A0A2L2YKP6 A0A026WNN1 A0A3L8DV95
Pubmed
19121390
22118469
26354079
26227816
17510324
12364791
+ More
14747013 17210077 26483478 20966253 24945155 25401762 26823975 26369729 17994087 22936249 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 26108605 20566863 25830018 24495485 15632085 23254933 24845553 26334808 27129103 28004739 25315136 25474469 26383154 17615350 18362917 19820115 26561354 24508170 30249741
14747013 17210077 26483478 20966253 24945155 25401762 26823975 26369729 17994087 22936249 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 26108605 20566863 25830018 24495485 15632085 23254933 24845553 26334808 27129103 28004739 25315136 25474469 26383154 17615350 18362917 19820115 26561354 24508170 30249741
EMBL
BABH01034496
ODYU01001422
SOQ37584.1
AGBW02014991
OWR40765.1
KQ458860
+ More
KPJ04776.1 KQ461191 KPJ07083.1 NWSH01002733 PCG67607.1 JTDY01002974 KOB70386.1 GFDF01003144 JAV10940.1 UFQT01000752 SSX26973.1 CH477228 EAT47071.1 AJVK01000592 AAAB01008944 EAA10204.2 DS231839 EDS34299.1 JXUM01079762 JXUM01079763 KQ563144 KXJ74398.1 GEDC01027500 GEDC01003066 GEDC01002257 JAS09798.1 JAS34232.1 JAS35041.1 GECZ01027562 JAS42207.1 GECU01022627 JAS85079.1 GAPW01004329 JAC09269.1 GFDL01013332 JAV21713.1 KK856646 PTY26174.1 APCN01000667 GBHO01000318 JAG43286.1 GBHO01000317 GBRD01008657 JAG43287.1 JAG57164.1 AXCM01014561 GBHO01002176 JAG41428.1 GDHC01015900 JAQ02729.1 GDAI01001989 JAI15614.1 GDHC01012740 JAQ05889.1 CCAG010023894 CM000361 CM002910 EDX04306.1 KMY89179.1 GAKP01002992 JAC55960.1 CH902620 EDV31846.1 CM000157 KRJ97923.1 CH480818 EDW52207.1 CH940654 EDW57924.2 AXCN02002186 AE014134 AAO41175.1 AHN54283.1 CH954177 EDV58310.1 JXJN01007520 PYGN01000083 PSN54797.1 GDHF01032662 JAI19652.1 JRES01001270 KNC24050.1 DS235843 EEB18282.1 GBXI01012001 JAD02291.1 CH479184 EDW37347.1 GAMC01003569 JAC02987.1 GECU01036453 GECU01009285 JAS71253.1 JAS98421.1 CH379062 EDY70710.1 CH916372 EDV99478.1 CP012523 ALC39014.1 CH963920 EDW77513.1 AMQN01008214 KB302448 ELU04327.1 KK852514 KDR22238.1 OUUW01000006 SPP81521.1 GBGD01002695 JAC86194.1 GDKW01001185 JAI55410.1 ACPB03008885 GAHY01001171 JAA76339.1 NEVH01002144 PNF42540.1 GGFM01008016 MBW28767.1 GFTR01004017 JAW12409.1 GEZM01101341 JAV52687.1 GBBI01001337 JAC17375.1 GGFJ01009389 MBW58530.1 CH933807 EDW11956.1 KQ982482 KYQ55920.1 GGFJ01009390 MBW58531.1 GGFL01004799 MBW68977.1 GGFJ01009391 MBW58532.1 AJWK01029092 AJWK01029093 GFDF01003234 JAV10850.1 KQ435696 KOX80826.1 GGFL01004642 MBW68820.1 GGFK01011845 MBW45166.1 HACG01016400 CEK63265.1 DS469777 EDO33325.1 KQ414716 KOC62799.1 KQ971343 EFA04414.1 IAAA01023060 IAAA01023061 IAAA01023062 IAAA01023063 LAA07925.1 KK107144 EZA57578.1 QOIP01000003 RLU24354.1
KPJ04776.1 KQ461191 KPJ07083.1 NWSH01002733 PCG67607.1 JTDY01002974 KOB70386.1 GFDF01003144 JAV10940.1 UFQT01000752 SSX26973.1 CH477228 EAT47071.1 AJVK01000592 AAAB01008944 EAA10204.2 DS231839 EDS34299.1 JXUM01079762 JXUM01079763 KQ563144 KXJ74398.1 GEDC01027500 GEDC01003066 GEDC01002257 JAS09798.1 JAS34232.1 JAS35041.1 GECZ01027562 JAS42207.1 GECU01022627 JAS85079.1 GAPW01004329 JAC09269.1 GFDL01013332 JAV21713.1 KK856646 PTY26174.1 APCN01000667 GBHO01000318 JAG43286.1 GBHO01000317 GBRD01008657 JAG43287.1 JAG57164.1 AXCM01014561 GBHO01002176 JAG41428.1 GDHC01015900 JAQ02729.1 GDAI01001989 JAI15614.1 GDHC01012740 JAQ05889.1 CCAG010023894 CM000361 CM002910 EDX04306.1 KMY89179.1 GAKP01002992 JAC55960.1 CH902620 EDV31846.1 CM000157 KRJ97923.1 CH480818 EDW52207.1 CH940654 EDW57924.2 AXCN02002186 AE014134 AAO41175.1 AHN54283.1 CH954177 EDV58310.1 JXJN01007520 PYGN01000083 PSN54797.1 GDHF01032662 JAI19652.1 JRES01001270 KNC24050.1 DS235843 EEB18282.1 GBXI01012001 JAD02291.1 CH479184 EDW37347.1 GAMC01003569 JAC02987.1 GECU01036453 GECU01009285 JAS71253.1 JAS98421.1 CH379062 EDY70710.1 CH916372 EDV99478.1 CP012523 ALC39014.1 CH963920 EDW77513.1 AMQN01008214 KB302448 ELU04327.1 KK852514 KDR22238.1 OUUW01000006 SPP81521.1 GBGD01002695 JAC86194.1 GDKW01001185 JAI55410.1 ACPB03008885 GAHY01001171 JAA76339.1 NEVH01002144 PNF42540.1 GGFM01008016 MBW28767.1 GFTR01004017 JAW12409.1 GEZM01101341 JAV52687.1 GBBI01001337 JAC17375.1 GGFJ01009389 MBW58530.1 CH933807 EDW11956.1 KQ982482 KYQ55920.1 GGFJ01009390 MBW58531.1 GGFL01004799 MBW68977.1 GGFJ01009391 MBW58532.1 AJWK01029092 AJWK01029093 GFDF01003234 JAV10850.1 KQ435696 KOX80826.1 GGFL01004642 MBW68820.1 GGFK01011845 MBW45166.1 HACG01016400 CEK63265.1 DS469777 EDO33325.1 KQ414716 KOC62799.1 KQ971343 EFA04414.1 IAAA01023060 IAAA01023061 IAAA01023062 IAAA01023063 LAA07925.1 KK107144 EZA57578.1 QOIP01000003 RLU24354.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000218220
UP000075901
+ More
UP000037510 UP000008820 UP000092462 UP000007062 UP000002320 UP000069940 UP000249989 UP000075902 UP000075882 UP000076407 UP000075903 UP000075900 UP000075840 UP000075920 UP000075883 UP000075881 UP000092444 UP000000304 UP000078200 UP000092445 UP000007801 UP000002282 UP000192221 UP000001292 UP000008792 UP000075886 UP000000803 UP000075884 UP000008711 UP000092460 UP000245037 UP000092443 UP000037069 UP000009046 UP000008744 UP000075880 UP000001819 UP000091820 UP000001070 UP000092553 UP000007798 UP000014760 UP000027135 UP000268350 UP000015103 UP000235965 UP000095301 UP000009192 UP000075809 UP000092461 UP000069272 UP000053105 UP000085678 UP000095300 UP000001593 UP000053825 UP000007266 UP000053097 UP000279307
UP000037510 UP000008820 UP000092462 UP000007062 UP000002320 UP000069940 UP000249989 UP000075902 UP000075882 UP000076407 UP000075903 UP000075900 UP000075840 UP000075920 UP000075883 UP000075881 UP000092444 UP000000304 UP000078200 UP000092445 UP000007801 UP000002282 UP000192221 UP000001292 UP000008792 UP000075886 UP000000803 UP000075884 UP000008711 UP000092460 UP000245037 UP000092443 UP000037069 UP000009046 UP000008744 UP000075880 UP000001819 UP000091820 UP000001070 UP000092553 UP000007798 UP000014760 UP000027135 UP000268350 UP000015103 UP000235965 UP000095301 UP000009192 UP000075809 UP000092461 UP000069272 UP000053105 UP000085678 UP000095300 UP000001593 UP000053825 UP000007266 UP000053097 UP000279307
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J2M5
A0A2H1VBH3
A0A212EGZ9
A0A194QMR5
A0A194QNI4
A0A2A4J666
+ More
A0A182STQ8 A0A0L7L4X7 A0A1L8DWU9 A0A336MDU9 Q17K21 A0A1B0DB08 Q7Q8B4 B0W4W1 A0A182GQ12 A0A182U7N4 A0A182KWE8 A0A182X6A6 A0A1B6EAU3 A0A1B6EW96 A0A1B6IDR8 A0A182V2H7 A0A023ELW7 A0A1Q3F2D0 A0A2R7X2Z2 A0A182R3Z6 A0A182IBC8 A0A182VTF8 A0A0A9ZGL6 A0A0A9ZDF2 A0A182MQE3 A0A0A9ZDL2 A0A146L4B1 A0A0K8TMQ0 A0A146LE30 A0A182K804 A0A1B0G0C5 B4Q7B2 A0A034WP43 A0A1A9VA62 A0A1B0AAK6 B3MM86 A0A0R1DSI4 A0A1W4VZN9 B4HYS7 B4M9P8 A0A182R0F7 Q86BM0 A0A182NAL7 B3N7U9 A0A1B0B291 A0A2P8ZE64 A0A1A9Y1P5 A0A0K8TYW1 A0A0L0BVG0 E0VY26 A0A0A1WUM9 B4GJ33 A0A182IQ93 W8C7K5 A0A1B6JGL8 B5DK84 A0A1A9W2J9 B4JP53 A0A0M4E9S4 B4N0A0 R7UM23 A0A067RGU5 A0A3B0K919 A0A069DXV5 A0A0P4W141 R4G421 A0A2J7RNX5 A0A2M3ZJK9 A0A224XWL3 A0A1Y1K1J4 A0A1I8N6M9 A0A023F7Z4 A0A2M4BZL7 B4KEY7 A0A151X6J4 A0A2M4C068 A0A2M4CUF7 A0A2M4BZS9 A0A1B0GKL3 A0A182F3C3 A0A1L8DWM6 A0A0M9AC56 A0A2M4CU21 A0A2M4AWI7 A0A0B6Z6B4 A0A1S3H8A0 A0A1I8Q4Y8 A7SSI3 A0A0L7QW63 D6WP02 A0A2L2YKP6 A0A026WNN1 A0A3L8DV95
A0A182STQ8 A0A0L7L4X7 A0A1L8DWU9 A0A336MDU9 Q17K21 A0A1B0DB08 Q7Q8B4 B0W4W1 A0A182GQ12 A0A182U7N4 A0A182KWE8 A0A182X6A6 A0A1B6EAU3 A0A1B6EW96 A0A1B6IDR8 A0A182V2H7 A0A023ELW7 A0A1Q3F2D0 A0A2R7X2Z2 A0A182R3Z6 A0A182IBC8 A0A182VTF8 A0A0A9ZGL6 A0A0A9ZDF2 A0A182MQE3 A0A0A9ZDL2 A0A146L4B1 A0A0K8TMQ0 A0A146LE30 A0A182K804 A0A1B0G0C5 B4Q7B2 A0A034WP43 A0A1A9VA62 A0A1B0AAK6 B3MM86 A0A0R1DSI4 A0A1W4VZN9 B4HYS7 B4M9P8 A0A182R0F7 Q86BM0 A0A182NAL7 B3N7U9 A0A1B0B291 A0A2P8ZE64 A0A1A9Y1P5 A0A0K8TYW1 A0A0L0BVG0 E0VY26 A0A0A1WUM9 B4GJ33 A0A182IQ93 W8C7K5 A0A1B6JGL8 B5DK84 A0A1A9W2J9 B4JP53 A0A0M4E9S4 B4N0A0 R7UM23 A0A067RGU5 A0A3B0K919 A0A069DXV5 A0A0P4W141 R4G421 A0A2J7RNX5 A0A2M3ZJK9 A0A224XWL3 A0A1Y1K1J4 A0A1I8N6M9 A0A023F7Z4 A0A2M4BZL7 B4KEY7 A0A151X6J4 A0A2M4C068 A0A2M4CUF7 A0A2M4BZS9 A0A1B0GKL3 A0A182F3C3 A0A1L8DWM6 A0A0M9AC56 A0A2M4CU21 A0A2M4AWI7 A0A0B6Z6B4 A0A1S3H8A0 A0A1I8Q4Y8 A7SSI3 A0A0L7QW63 D6WP02 A0A2L2YKP6 A0A026WNN1 A0A3L8DV95
PDB
2P5X
E-value=1.20656e-44,
Score=449
Ontologies
GO
PANTHER
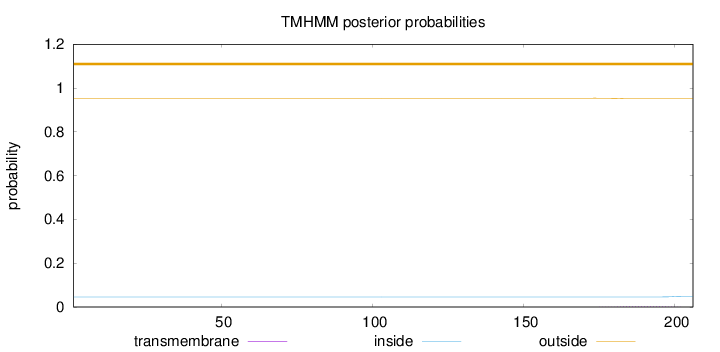
Topology
Length:
206
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05211
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04623
outside
1 - 206
Population Genetic Test Statistics
Pi
17.407085
Theta
17.626584
Tajima's D
-0.308139
CLR
1.080287
CSRT
0.291785410729464
Interpretation
Uncertain
