Gene
KWMTBOMO02319
Annotation
PREDICTED:_uncharacterized_protein_LOC105842814?_partial_[Bombyx_mori]
Full name
Peptidylprolyl isomerase
Location in the cell
Nuclear Reliability : 2.351
Sequence
CDS
ATGGCTGAGTGGAATGAAGACACTGATTCTGATATTGAAAATTTAGAAATTCTCAATGACTTCAATTCGTATTTTAACAGACCTTATCTAGTCAGACCAAGAGAAAACAATTTCGTTAAGTGGGACGATACGGAGTTTATACGTCGTTTTCGTGTATCTAAGAATACAGCTTTTACGATTTTAGAAGAAATCGCTAATGACATATCACACCCCACAAATCGGAATCATGCATTTTCAGCGTATAACCAGTTATTATTAACCTTGGCTACTGGATGCATGCAAATTACTGTTGGCGATTTCAGTGGTGTTCACCAAACGACAGCAGGAAAGGCTATTAAAAAAGTTACCGAAGCGATTGCATCAAGACGATGCCAATATATAATGCTGTAA
Protein
MAEWNEDTDSDIENLEILNDFNSYFNRPYLVRPRENNFVKWDDTEFIRRFRVSKNTAFTILEEIANDISHPTNRNHAFSAYNQLLLTLATGCMQITVGDFSGVHQTTAGKAIKKVTEAIASRRCQYIML
Summary
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Uniprot
X1XI09
J9LE40
A0A1Y1MC35
E9J220
A0A151IFZ5
A0A2S2N9H9
+ More
A0A1Y1KRW4 C4WUD0 A0A0L7KSD9 J9M2N2 A0A0A9X6A9 A0A2S2PS38 J9LYS1 J9LXS6 J9JZ87 U4UPE5 A0A0J7JXA2 U4U2V2 A0A1Y1KQW0 A0A151J9H7 A0A2S2NS44 J9L0T4 J9LUG4 J9LL64 J9M2S7 V5IB13 X1X0S1 A0A2S2NKK2 A0A151J5X9 A0A151I717 A0A151IQQ8 J9M943 J9JTM0 A0A1Y1M7D0 A0A1Y1MBC4 J9K8M3 N6TRS6 J9JJW2 A0A2P8Z014 E2B6X1 J9KWE0 A0A0A9ZIH6 A0A1Y1MXR4 X1XA32 A0A0L7LT10 A0A1Y1MZ45 A0A2S2NIF8 J9KK24 U4ULG9 J9L523 J9KYE0 J9KLH0 A0A151I7M4 A0A0L7KN33 A0A3Q0J0W5 A0A0L7KYL1 N6UD62 J9JXP4 J9L1M2 X1WR39 J9M4N8 V5G8D2 U4UCR2 J9LUU4 A0A151I6Z5 J9JWW4 C4WVD2 H9JFA0 A0A1Y1K586 J9KI14 S4PD89 A0A1I8NJZ2 A0A2P8XEM7 X1XEZ0 A0A151WZD3 A0A0L7L6V8 A0A0L7L7T8 A0A0L7L8T7 J9JM90 J9L1W5 A0A1Y1KLA7 A0A0L7L248 A0A1S3J8B0 W4YUY4 A0A0L7KN79 A0A1I8NIS5 J9LL02 X1WYW9 D6WYB2 A0A2R2MN13 A0A1S3JPJ3 J9LEH9 A0A147BLX3
A0A1Y1KRW4 C4WUD0 A0A0L7KSD9 J9M2N2 A0A0A9X6A9 A0A2S2PS38 J9LYS1 J9LXS6 J9JZ87 U4UPE5 A0A0J7JXA2 U4U2V2 A0A1Y1KQW0 A0A151J9H7 A0A2S2NS44 J9L0T4 J9LUG4 J9LL64 J9M2S7 V5IB13 X1X0S1 A0A2S2NKK2 A0A151J5X9 A0A151I717 A0A151IQQ8 J9M943 J9JTM0 A0A1Y1M7D0 A0A1Y1MBC4 J9K8M3 N6TRS6 J9JJW2 A0A2P8Z014 E2B6X1 J9KWE0 A0A0A9ZIH6 A0A1Y1MXR4 X1XA32 A0A0L7LT10 A0A1Y1MZ45 A0A2S2NIF8 J9KK24 U4ULG9 J9L523 J9KYE0 J9KLH0 A0A151I7M4 A0A0L7KN33 A0A3Q0J0W5 A0A0L7KYL1 N6UD62 J9JXP4 J9L1M2 X1WR39 J9M4N8 V5G8D2 U4UCR2 J9LUU4 A0A151I6Z5 J9JWW4 C4WVD2 H9JFA0 A0A1Y1K586 J9KI14 S4PD89 A0A1I8NJZ2 A0A2P8XEM7 X1XEZ0 A0A151WZD3 A0A0L7L6V8 A0A0L7L7T8 A0A0L7L8T7 J9JM90 J9L1W5 A0A1Y1KLA7 A0A0L7L248 A0A1S3J8B0 W4YUY4 A0A0L7KN79 A0A1I8NIS5 J9LL02 X1WYW9 D6WYB2 A0A2R2MN13 A0A1S3JPJ3 J9LEH9 A0A147BLX3
EC Number
5.2.1.8
Pubmed
EMBL
ABLF02012281
ABLF02058185
ABLF02031502
ABLF02037237
GEZM01035464
GEZM01035462
+ More
JAV83213.1 GL767726 EFZ13133.1 KQ977745 KYN00155.1 GGMR01000787 MBY13406.1 GEZM01078171 GEZM01078169 GEZM01078168 GEZM01078162 GEZM01078161 GEZM01078158 GEZM01078157 GEZM01078155 GEZM01078153 GEZM01078152 GEZM01078149 GEZM01078144 GEZM01078140 GEZM01078137 JAV62920.1 ABLF02006541 ABLF02042859 AK341014 BAH71500.1 JTDY01006499 KOB65959.1 ABLF02020885 GBHO01029241 JAG14363.1 GGMR01019661 MBY32280.1 ABLF02012895 ABLF02003846 ABLF02036739 KI208887 ERL95949.1 LBMM01023459 KMQ82717.1 KB631844 ERL86663.1 GEZM01076717 JAV63704.1 KQ979429 KYN21603.1 GGMR01007355 MBY19974.1 ABLF02003216 ABLF02024993 ABLF02024998 ABLF02025000 ABLF02019988 ABLF02066411 ABLF02011053 GALX01000235 JAB68231.1 ABLF02020738 GGMR01005102 MBY17721.1 KQ979914 KYN18634.1 KQ978474 KYM93682.1 KQ976758 KYN08556.1 ABLF02017542 ABLF02029655 ABLF02030019 GEZM01038551 JAV81614.1 GEZM01038550 JAV81615.1 ABLF02006612 APGK01006027 KB736544 ENN83189.1 ABLF02011428 PYGN01000260 PSN49845.1 GL446051 EFN88560.1 ABLF02016012 ABLF02055237 GBHO01000839 JAG42765.1 GEZM01017848 JAV90419.1 ABLF02021397 JTDY01000151 KOB78582.1 GEZM01017851 GEZM01017841 GEZM01017840 JAV90428.1 GGMR01004348 MBY16967.1 ABLF02038239 KB632267 ERL91031.1 ABLF02035722 ABLF02019697 ABLF02022234 ABLF02058482 KQ978403 KYM94175.1 JTDY01008190 KOB64687.1 JTDY01004268 KOB68358.1 APGK01040149 KB740975 ENN76582.1 ABLF02008545 ABLF02031609 ABLF02063305 ABLF02015087 GALX01002086 JAB66380.1 KB632204 ERL90113.1 ABLF02007356 KQ978464 KYM93783.1 ABLF02011552 AK341464 BAH71852.1 BABH01027007 GEZM01092234 JAV56642.1 ABLF02015760 GAIX01003761 JAA88799.1 PYGN01002465 PSN30448.1 ABLF02062536 KQ982919 KQ982640 KYQ49272.1 KYQ53248.1 JTDY01002554 KOB71187.1 JTDY01002465 KOB71356.1 JTDY01002286 KOB71739.1 ABLF02016431 ABLF02019815 ABLF02019816 GEZM01080383 JAV62179.1 JTDY01003471 KOB69505.1 AAGJ04098203 JTDY01008717 KOB64434.1 ABLF02038506 ABLF02016048 KQ971356 EFA09128.1 ABLF02022956 ABLF02059298 ABLF02064425 GEGO01003648 JAR91756.1
JAV83213.1 GL767726 EFZ13133.1 KQ977745 KYN00155.1 GGMR01000787 MBY13406.1 GEZM01078171 GEZM01078169 GEZM01078168 GEZM01078162 GEZM01078161 GEZM01078158 GEZM01078157 GEZM01078155 GEZM01078153 GEZM01078152 GEZM01078149 GEZM01078144 GEZM01078140 GEZM01078137 JAV62920.1 ABLF02006541 ABLF02042859 AK341014 BAH71500.1 JTDY01006499 KOB65959.1 ABLF02020885 GBHO01029241 JAG14363.1 GGMR01019661 MBY32280.1 ABLF02012895 ABLF02003846 ABLF02036739 KI208887 ERL95949.1 LBMM01023459 KMQ82717.1 KB631844 ERL86663.1 GEZM01076717 JAV63704.1 KQ979429 KYN21603.1 GGMR01007355 MBY19974.1 ABLF02003216 ABLF02024993 ABLF02024998 ABLF02025000 ABLF02019988 ABLF02066411 ABLF02011053 GALX01000235 JAB68231.1 ABLF02020738 GGMR01005102 MBY17721.1 KQ979914 KYN18634.1 KQ978474 KYM93682.1 KQ976758 KYN08556.1 ABLF02017542 ABLF02029655 ABLF02030019 GEZM01038551 JAV81614.1 GEZM01038550 JAV81615.1 ABLF02006612 APGK01006027 KB736544 ENN83189.1 ABLF02011428 PYGN01000260 PSN49845.1 GL446051 EFN88560.1 ABLF02016012 ABLF02055237 GBHO01000839 JAG42765.1 GEZM01017848 JAV90419.1 ABLF02021397 JTDY01000151 KOB78582.1 GEZM01017851 GEZM01017841 GEZM01017840 JAV90428.1 GGMR01004348 MBY16967.1 ABLF02038239 KB632267 ERL91031.1 ABLF02035722 ABLF02019697 ABLF02022234 ABLF02058482 KQ978403 KYM94175.1 JTDY01008190 KOB64687.1 JTDY01004268 KOB68358.1 APGK01040149 KB740975 ENN76582.1 ABLF02008545 ABLF02031609 ABLF02063305 ABLF02015087 GALX01002086 JAB66380.1 KB632204 ERL90113.1 ABLF02007356 KQ978464 KYM93783.1 ABLF02011552 AK341464 BAH71852.1 BABH01027007 GEZM01092234 JAV56642.1 ABLF02015760 GAIX01003761 JAA88799.1 PYGN01002465 PSN30448.1 ABLF02062536 KQ982919 KQ982640 KYQ49272.1 KYQ53248.1 JTDY01002554 KOB71187.1 JTDY01002465 KOB71356.1 JTDY01002286 KOB71739.1 ABLF02016431 ABLF02019815 ABLF02019816 GEZM01080383 JAV62179.1 JTDY01003471 KOB69505.1 AAGJ04098203 JTDY01008717 KOB64434.1 ABLF02038506 ABLF02016048 KQ971356 EFA09128.1 ABLF02022956 ABLF02059298 ABLF02064425 GEGO01003648 JAR91756.1
Proteomes
PRIDE
Pfam
Interpro
SUPFAM
SSF53098
SSF53098
ProteinModelPortal
X1XI09
J9LE40
A0A1Y1MC35
E9J220
A0A151IFZ5
A0A2S2N9H9
+ More
A0A1Y1KRW4 C4WUD0 A0A0L7KSD9 J9M2N2 A0A0A9X6A9 A0A2S2PS38 J9LYS1 J9LXS6 J9JZ87 U4UPE5 A0A0J7JXA2 U4U2V2 A0A1Y1KQW0 A0A151J9H7 A0A2S2NS44 J9L0T4 J9LUG4 J9LL64 J9M2S7 V5IB13 X1X0S1 A0A2S2NKK2 A0A151J5X9 A0A151I717 A0A151IQQ8 J9M943 J9JTM0 A0A1Y1M7D0 A0A1Y1MBC4 J9K8M3 N6TRS6 J9JJW2 A0A2P8Z014 E2B6X1 J9KWE0 A0A0A9ZIH6 A0A1Y1MXR4 X1XA32 A0A0L7LT10 A0A1Y1MZ45 A0A2S2NIF8 J9KK24 U4ULG9 J9L523 J9KYE0 J9KLH0 A0A151I7M4 A0A0L7KN33 A0A3Q0J0W5 A0A0L7KYL1 N6UD62 J9JXP4 J9L1M2 X1WR39 J9M4N8 V5G8D2 U4UCR2 J9LUU4 A0A151I6Z5 J9JWW4 C4WVD2 H9JFA0 A0A1Y1K586 J9KI14 S4PD89 A0A1I8NJZ2 A0A2P8XEM7 X1XEZ0 A0A151WZD3 A0A0L7L6V8 A0A0L7L7T8 A0A0L7L8T7 J9JM90 J9L1W5 A0A1Y1KLA7 A0A0L7L248 A0A1S3J8B0 W4YUY4 A0A0L7KN79 A0A1I8NIS5 J9LL02 X1WYW9 D6WYB2 A0A2R2MN13 A0A1S3JPJ3 J9LEH9 A0A147BLX3
A0A1Y1KRW4 C4WUD0 A0A0L7KSD9 J9M2N2 A0A0A9X6A9 A0A2S2PS38 J9LYS1 J9LXS6 J9JZ87 U4UPE5 A0A0J7JXA2 U4U2V2 A0A1Y1KQW0 A0A151J9H7 A0A2S2NS44 J9L0T4 J9LUG4 J9LL64 J9M2S7 V5IB13 X1X0S1 A0A2S2NKK2 A0A151J5X9 A0A151I717 A0A151IQQ8 J9M943 J9JTM0 A0A1Y1M7D0 A0A1Y1MBC4 J9K8M3 N6TRS6 J9JJW2 A0A2P8Z014 E2B6X1 J9KWE0 A0A0A9ZIH6 A0A1Y1MXR4 X1XA32 A0A0L7LT10 A0A1Y1MZ45 A0A2S2NIF8 J9KK24 U4ULG9 J9L523 J9KYE0 J9KLH0 A0A151I7M4 A0A0L7KN33 A0A3Q0J0W5 A0A0L7KYL1 N6UD62 J9JXP4 J9L1M2 X1WR39 J9M4N8 V5G8D2 U4UCR2 J9LUU4 A0A151I6Z5 J9JWW4 C4WVD2 H9JFA0 A0A1Y1K586 J9KI14 S4PD89 A0A1I8NJZ2 A0A2P8XEM7 X1XEZ0 A0A151WZD3 A0A0L7L6V8 A0A0L7L7T8 A0A0L7L8T7 J9JM90 J9L1W5 A0A1Y1KLA7 A0A0L7L248 A0A1S3J8B0 W4YUY4 A0A0L7KN79 A0A1I8NIS5 J9LL02 X1WYW9 D6WYB2 A0A2R2MN13 A0A1S3JPJ3 J9LEH9 A0A147BLX3
Ontologies
PANTHER
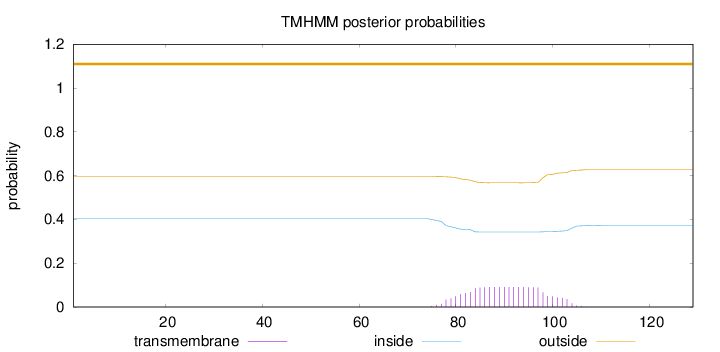
Topology
Length:
129
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.90913
Exp number, first 60 AAs:
0.00025
Total prob of N-in:
0.40346
outside
1 - 129
Population Genetic Test Statistics
Pi
8.476696
Theta
16.023184
Tajima's D
-1.523621
CLR
85.196595
CSRT
0.0550972451377431
Interpretation
Uncertain
