Gene
KWMTBOMO02317
Pre Gene Modal
BGIBMGA003786
Annotation
PREDICTED:_voltage-dependent_L-type_calcium_channel_subunit_beta-2_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.077
Sequence
CDS
ATGTGTTTGTTTGTTGTTTTCCAGGGCTCCGCGGAGTCGAATTGTTCGCAGCCGTCGTCGGAGCTGTCGCTGGACGATGACAAGGAGGCGCTGCGCAGAGAAAAGGAAGCGCAGGCTCTCTCACAGCTGGATAAGGCGAGGTCAAAGCCGGTCGCGTTCGCCGTCCGCACGAACGTCGCTTACGACGCGAAGATCGACGACGACTCGCCCGTGCACGGCACCGCCATCTCCTTCGACGTTCGAGACTTCCTGCACATTAAGGAGAAGTACGACAACAACTGGTGGATCGGTCGGCTCGTGGCGGAGAACTCTGACATAGGGTTCATCCCGTCCCCCGTGAAGCTGGAGATGGTGCGCGCGGGGCTGGCCGCCAGGGGCCGCTCCGCGCTCGCCGCCCACCCCCACTCCCGAGGTAGCACTCCCCCCACGCCCGGTGACGAATCCGACTCGGCGGGCAGGGGGCGCGGGGCGGGCGCGCTGCCCGCTGGCAAGGAAAAGAAGAAGCCATTCTTCAAGAAGCAGGAGGCCTGCACTCCGTACGACGTGGTGCCCTCGATGAGGCCGGTCGTCCTCGTGGGGCCCTCGCTCAAAGGATACGAAGTCACGGACATGATGCAGAAGGCGCTTTTTGATTTCTTAAAAAGGCGCTTCGAAGGACGGATAATAATAACGAGGGTGATGGCGGACATATCGCTGGCGAAGCGCTCGCTGCTCAACAACCCCTCCAAGAGGGCCATCATGGAGCGCTCCAACTCGCGCTCCACCTGCCTGGCCGAGGTGCAGGCTGAAATCGAGAGGATATTCGACCTAGCGCGGACCCTGCAGCTGGTGGTGCTGGACTGCGACACCATCAACCACCCGTCGCAACTGGCCAAGACCTCACTAGCGCCCACTATCGTCTACTTAAAGATCACCAGCCCCAAGGTGCTGCAGCGATTGATCAAGTCCCGGGGCAAGGGCCAGACCAAGAACCTGTCCGTGCAAATGGTGGCGGCCGAGAAGCTGTCGCAGTGTCCCAAGGAGATGTTCGACCTCGTGCTGGACGAAAACCAGCTGGAGGACGCCTGCGACCGCATCGCAGAGCATCTGGAGAAGTACTGGCGCGCCACGCACCCCCCAGCCGCCGCCCCGCTGGCGCCGCCCGCCGCCCCCCGCGCGCACCTCGCCCCCCTGCCCCACCTCGCGCACCTCGCGCACGCCCAGCCCGTACAAGCGTCTGGCGGGCGAGCCCGCGGGCCGCGCCCCGAGCGGCTGGAGGAGGAGTACTACGGGCGGGAGCGGTACGCGCGCGAGTACGAGTACGGACACGACTACACGCGCCGCGACCCCGACGCCGACGAGTACGGGCCCTCGCGCCGAGCGCTCAACGCTGTCTAG
Protein
MCLFVVFQGSAESNCSQPSSELSLDDDKEALRREKEAQALSQLDKARSKPVAFAVRTNVAYDAKIDDDSPVHGTAISFDVRDFLHIKEKYDNNWWIGRLVAENSDIGFIPSPVKLEMVRAGLAARGRSALAAHPHSRGSTPPTPGDESDSAGRGRGAGALPAGKEKKKPFFKKQEACTPYDVVPSMRPVVLVGPSLKGYEVTDMMQKALFDFLKRRFEGRIIITRVMADISLAKRSLLNNPSKRAIMERSNSRSTCLAEVQAEIERIFDLARTLQLVVLDCDTINHPSQLAKTSLAPTIVYLKITSPKVLQRLIKSRGKGQTKNLSVQMVAAEKLSQCPKEMFDLVLDENQLEDACDRIAEHLEKYWRATHPPAAAPLAPPAAPRAHLAPLPHLAHLAHAQPVQASGGRARGPRPERLEEEYYGRERYAREYEYGHDYTRRDPDADEYGPSRRALNAV
Summary
Uniprot
A0A2A4JPC1
A0A2A4JP69
A0A212EML9
A0A2A4JNF4
A0A194QH78
V9IAN0
+ More
A0A087ZMZ3 A0A1Y1KBK3 A0A1Y1K7U2 A0A1Y1K818 A0A1Y1KFL1 A0A1Y1KD44 A0A1Y1K833 T1SG89 A0A1Y1KFK1 A0A1Y1KDR8 A0A1Y1K802 A0A139WHQ7 A0A1B0CUU4 A0A139WHE7 A0A1I8PM22 A0A026W8H4 E2ASH3 Q7M3V8 A0A154P7Q5 A0A0A1XKZ5 K7INI2 A0A195D0B8 A0A034WNH0 A0A034WNG5 A0A0K8TXP2 A0A034WLT2 A0A336N317 A0A336MYH7 A0A0Q9X6P7 A0A0K8WDX8 A0A0R1DKW6 N6TM54 A0A0Q9WJQ1 W8BJL5 A0A034WIC8 A0A1W4W4Z8 A0A1W4W449 A0A0J9R0P5 A0A195D8K3 Q7KTE8 A0A034WMC0 A0A0Q5WHL4 A0A151WYC9 A0A0J9R089 A0A0Q9X637 A0A0R3NSR7 A0A0Q9W8L1 M9PD84 A0A0Q5WAT6 W8CB80 A0A1B0G029 A0A195B8Z5 A0A0R1DSD5 A0A0Q9XD98 V9IA55 T1SG87 A0A1W4VSX4 W8BJL9 A0A1A9XE24 A0A0Q9WJJ8 A0A3B0JAW8 B4MWD8 A0A2A4JPH6 W8C560 A0A2A4JQC5 T1SG86 A0A1B0A0R7 A0A1A9UIL1 A0A1I8PLY9 A0A1I8PLX9 A0A1I8PM12 A0A1I8PLU1 A0A1I8PLY4 A0A1I8PM04 A0A1W4VSU5 A0A0Q9W8N5 A0A0R3NTE8 T1PHY8 A0A2A4JNJ5 A0A0Q5WA93 A0A0J9R1K8 M9PFL4 A0A0Q5WJW1 B3N4K0 A0A0R3NTM1 A0A0R3NTB1 A0A0Q9WIH9 B4KHA4 A0A0Q5WNZ7 Q29M31 A0A034WKZ1 A0A1I8PLV3 A0A1Q3FZK5 A0A0J9R1L8
A0A087ZMZ3 A0A1Y1KBK3 A0A1Y1K7U2 A0A1Y1K818 A0A1Y1KFL1 A0A1Y1KD44 A0A1Y1K833 T1SG89 A0A1Y1KFK1 A0A1Y1KDR8 A0A1Y1K802 A0A139WHQ7 A0A1B0CUU4 A0A139WHE7 A0A1I8PM22 A0A026W8H4 E2ASH3 Q7M3V8 A0A154P7Q5 A0A0A1XKZ5 K7INI2 A0A195D0B8 A0A034WNH0 A0A034WNG5 A0A0K8TXP2 A0A034WLT2 A0A336N317 A0A336MYH7 A0A0Q9X6P7 A0A0K8WDX8 A0A0R1DKW6 N6TM54 A0A0Q9WJQ1 W8BJL5 A0A034WIC8 A0A1W4W4Z8 A0A1W4W449 A0A0J9R0P5 A0A195D8K3 Q7KTE8 A0A034WMC0 A0A0Q5WHL4 A0A151WYC9 A0A0J9R089 A0A0Q9X637 A0A0R3NSR7 A0A0Q9W8L1 M9PD84 A0A0Q5WAT6 W8CB80 A0A1B0G029 A0A195B8Z5 A0A0R1DSD5 A0A0Q9XD98 V9IA55 T1SG87 A0A1W4VSX4 W8BJL9 A0A1A9XE24 A0A0Q9WJJ8 A0A3B0JAW8 B4MWD8 A0A2A4JPH6 W8C560 A0A2A4JQC5 T1SG86 A0A1B0A0R7 A0A1A9UIL1 A0A1I8PLY9 A0A1I8PLX9 A0A1I8PM12 A0A1I8PLU1 A0A1I8PLY4 A0A1I8PM04 A0A1W4VSU5 A0A0Q9W8N5 A0A0R3NTE8 T1PHY8 A0A2A4JNJ5 A0A0Q5WA93 A0A0J9R1K8 M9PFL4 A0A0Q5WJW1 B3N4K0 A0A0R3NTM1 A0A0R3NTB1 A0A0Q9WIH9 B4KHA4 A0A0Q5WNZ7 Q29M31 A0A034WKZ1 A0A1I8PLV3 A0A1Q3FZK5 A0A0J9R1L8
Pubmed
EMBL
NWSH01000903
PCG73606.1
PCG73609.1
AGBW02013812
OWR42733.1
PCG73605.1
+ More
KQ458860 KPJ04774.1 JR037014 AEY57526.1 GEZM01089342 GEZM01089340 JAV57540.1 GEZM01089350 JAV57532.1 GEZM01089348 JAV57534.1 GEZM01089339 GEZM01089332 JAV57547.1 GEZM01089347 JAV57535.1 GEZM01089336 JAV57544.1 JX997993 AGT38440.1 GEZM01089344 JAV57537.1 GEZM01089335 GEZM01089334 JAV57546.1 GEZM01089349 JAV57533.1 KQ971343 KYB27419.1 AJWK01029657 AJWK01029658 KYB27418.1 KK107347 EZA52355.1 GL442298 EFN63639.1 KQ434822 KZC07160.1 GBXI01010712 GBXI01002732 JAD03580.1 JAD11560.1 KQ977012 KYN06363.1 GAKP01003634 JAC55318.1 GAKP01003639 GAKP01003637 GAKP01003635 JAC55313.1 GDHF01033253 GDHF01010658 JAI19061.1 JAI41656.1 GAKP01003640 JAC55312.1 UFQT01004028 SSX35443.1 SSX35442.1 CH933807 KRG03693.1 GDHF01003080 JAI49234.1 CM000157 KRJ97550.1 APGK01018891 KB740085 KB632303 ENN81534.1 ERL91800.1 CH940649 KRF81099.1 GAMC01005085 JAC01471.1 GAKP01003641 GAKP01003638 JAC55311.1 CM002910 KMY89718.1 KQ981135 KYN09213.1 AE014134 AAS64679.3 GAKP01003642 JAC55310.1 CH954177 KQS70565.1 KQ982650 KYQ52894.1 KMY89722.1 KRG03695.1 CH379060 KRT04273.1 KRF81101.1 AGB92910.1 KQS70566.1 GAMC01005081 JAC01475.1 CCAG010019099 KQ976558 KYM80700.1 KRJ97542.1 KRG03690.1 JR037015 AEY57527.1 JX997992 AGT38439.1 GAMC01005080 JAC01476.1 KRF81102.1 OUUW01000004 SPP79487.1 CH963857 EDW76008.2 PCG73608.1 GAMC01005084 JAC01472.1 PCG73610.1 JX997991 AGT38438.1 KRF81100.1 KRT04281.1 KA648386 AFP63015.1 PCG73607.1 KQS70567.1 KMY89724.1 AGB92912.1 KQS70569.1 EDV58912.2 KRT04284.1 KRT04276.1 KRF81103.1 EDW13321.2 KQS70568.1 EAL33864.3 GAKP01003633 JAC55319.1 GFDL01002050 JAV32995.1 KMY89734.1
KQ458860 KPJ04774.1 JR037014 AEY57526.1 GEZM01089342 GEZM01089340 JAV57540.1 GEZM01089350 JAV57532.1 GEZM01089348 JAV57534.1 GEZM01089339 GEZM01089332 JAV57547.1 GEZM01089347 JAV57535.1 GEZM01089336 JAV57544.1 JX997993 AGT38440.1 GEZM01089344 JAV57537.1 GEZM01089335 GEZM01089334 JAV57546.1 GEZM01089349 JAV57533.1 KQ971343 KYB27419.1 AJWK01029657 AJWK01029658 KYB27418.1 KK107347 EZA52355.1 GL442298 EFN63639.1 KQ434822 KZC07160.1 GBXI01010712 GBXI01002732 JAD03580.1 JAD11560.1 KQ977012 KYN06363.1 GAKP01003634 JAC55318.1 GAKP01003639 GAKP01003637 GAKP01003635 JAC55313.1 GDHF01033253 GDHF01010658 JAI19061.1 JAI41656.1 GAKP01003640 JAC55312.1 UFQT01004028 SSX35443.1 SSX35442.1 CH933807 KRG03693.1 GDHF01003080 JAI49234.1 CM000157 KRJ97550.1 APGK01018891 KB740085 KB632303 ENN81534.1 ERL91800.1 CH940649 KRF81099.1 GAMC01005085 JAC01471.1 GAKP01003641 GAKP01003638 JAC55311.1 CM002910 KMY89718.1 KQ981135 KYN09213.1 AE014134 AAS64679.3 GAKP01003642 JAC55310.1 CH954177 KQS70565.1 KQ982650 KYQ52894.1 KMY89722.1 KRG03695.1 CH379060 KRT04273.1 KRF81101.1 AGB92910.1 KQS70566.1 GAMC01005081 JAC01475.1 CCAG010019099 KQ976558 KYM80700.1 KRJ97542.1 KRG03690.1 JR037015 AEY57527.1 JX997992 AGT38439.1 GAMC01005080 JAC01476.1 KRF81102.1 OUUW01000004 SPP79487.1 CH963857 EDW76008.2 PCG73608.1 GAMC01005084 JAC01472.1 PCG73610.1 JX997991 AGT38438.1 KRF81100.1 KRT04281.1 KA648386 AFP63015.1 PCG73607.1 KQS70567.1 KMY89724.1 AGB92912.1 KQS70569.1 EDV58912.2 KRT04284.1 KRT04276.1 KRF81103.1 EDW13321.2 KQS70568.1 EAL33864.3 GAKP01003633 JAC55319.1 GFDL01002050 JAV32995.1 KMY89734.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000005203
UP000007266
UP000092461
+ More
UP000095300 UP000053097 UP000000311 UP000076502 UP000002358 UP000078542 UP000009192 UP000002282 UP000019118 UP000030742 UP000008792 UP000192221 UP000078492 UP000000803 UP000008711 UP000075809 UP000001819 UP000092444 UP000078540 UP000092443 UP000268350 UP000007798 UP000092445 UP000078200
UP000095300 UP000053097 UP000000311 UP000076502 UP000002358 UP000078542 UP000009192 UP000002282 UP000019118 UP000030742 UP000008792 UP000192221 UP000078492 UP000000803 UP000008711 UP000075809 UP000001819 UP000092444 UP000078540 UP000092443 UP000268350 UP000007798 UP000092445 UP000078200
PRIDE
Interpro
ProteinModelPortal
A0A2A4JPC1
A0A2A4JP69
A0A212EML9
A0A2A4JNF4
A0A194QH78
V9IAN0
+ More
A0A087ZMZ3 A0A1Y1KBK3 A0A1Y1K7U2 A0A1Y1K818 A0A1Y1KFL1 A0A1Y1KD44 A0A1Y1K833 T1SG89 A0A1Y1KFK1 A0A1Y1KDR8 A0A1Y1K802 A0A139WHQ7 A0A1B0CUU4 A0A139WHE7 A0A1I8PM22 A0A026W8H4 E2ASH3 Q7M3V8 A0A154P7Q5 A0A0A1XKZ5 K7INI2 A0A195D0B8 A0A034WNH0 A0A034WNG5 A0A0K8TXP2 A0A034WLT2 A0A336N317 A0A336MYH7 A0A0Q9X6P7 A0A0K8WDX8 A0A0R1DKW6 N6TM54 A0A0Q9WJQ1 W8BJL5 A0A034WIC8 A0A1W4W4Z8 A0A1W4W449 A0A0J9R0P5 A0A195D8K3 Q7KTE8 A0A034WMC0 A0A0Q5WHL4 A0A151WYC9 A0A0J9R089 A0A0Q9X637 A0A0R3NSR7 A0A0Q9W8L1 M9PD84 A0A0Q5WAT6 W8CB80 A0A1B0G029 A0A195B8Z5 A0A0R1DSD5 A0A0Q9XD98 V9IA55 T1SG87 A0A1W4VSX4 W8BJL9 A0A1A9XE24 A0A0Q9WJJ8 A0A3B0JAW8 B4MWD8 A0A2A4JPH6 W8C560 A0A2A4JQC5 T1SG86 A0A1B0A0R7 A0A1A9UIL1 A0A1I8PLY9 A0A1I8PLX9 A0A1I8PM12 A0A1I8PLU1 A0A1I8PLY4 A0A1I8PM04 A0A1W4VSU5 A0A0Q9W8N5 A0A0R3NTE8 T1PHY8 A0A2A4JNJ5 A0A0Q5WA93 A0A0J9R1K8 M9PFL4 A0A0Q5WJW1 B3N4K0 A0A0R3NTM1 A0A0R3NTB1 A0A0Q9WIH9 B4KHA4 A0A0Q5WNZ7 Q29M31 A0A034WKZ1 A0A1I8PLV3 A0A1Q3FZK5 A0A0J9R1L8
A0A087ZMZ3 A0A1Y1KBK3 A0A1Y1K7U2 A0A1Y1K818 A0A1Y1KFL1 A0A1Y1KD44 A0A1Y1K833 T1SG89 A0A1Y1KFK1 A0A1Y1KDR8 A0A1Y1K802 A0A139WHQ7 A0A1B0CUU4 A0A139WHE7 A0A1I8PM22 A0A026W8H4 E2ASH3 Q7M3V8 A0A154P7Q5 A0A0A1XKZ5 K7INI2 A0A195D0B8 A0A034WNH0 A0A034WNG5 A0A0K8TXP2 A0A034WLT2 A0A336N317 A0A336MYH7 A0A0Q9X6P7 A0A0K8WDX8 A0A0R1DKW6 N6TM54 A0A0Q9WJQ1 W8BJL5 A0A034WIC8 A0A1W4W4Z8 A0A1W4W449 A0A0J9R0P5 A0A195D8K3 Q7KTE8 A0A034WMC0 A0A0Q5WHL4 A0A151WYC9 A0A0J9R089 A0A0Q9X637 A0A0R3NSR7 A0A0Q9W8L1 M9PD84 A0A0Q5WAT6 W8CB80 A0A1B0G029 A0A195B8Z5 A0A0R1DSD5 A0A0Q9XD98 V9IA55 T1SG87 A0A1W4VSX4 W8BJL9 A0A1A9XE24 A0A0Q9WJJ8 A0A3B0JAW8 B4MWD8 A0A2A4JPH6 W8C560 A0A2A4JQC5 T1SG86 A0A1B0A0R7 A0A1A9UIL1 A0A1I8PLY9 A0A1I8PLX9 A0A1I8PM12 A0A1I8PLU1 A0A1I8PLY4 A0A1I8PM04 A0A1W4VSU5 A0A0Q9W8N5 A0A0R3NTE8 T1PHY8 A0A2A4JNJ5 A0A0Q5WA93 A0A0J9R1K8 M9PFL4 A0A0Q5WJW1 B3N4K0 A0A0R3NTM1 A0A0R3NTB1 A0A0Q9WIH9 B4KHA4 A0A0Q5WNZ7 Q29M31 A0A034WKZ1 A0A1I8PLV3 A0A1Q3FZK5 A0A0J9R1L8
PDB
1VYV
E-value=6.30308e-118,
Score=1086
Ontologies
GO
Topology
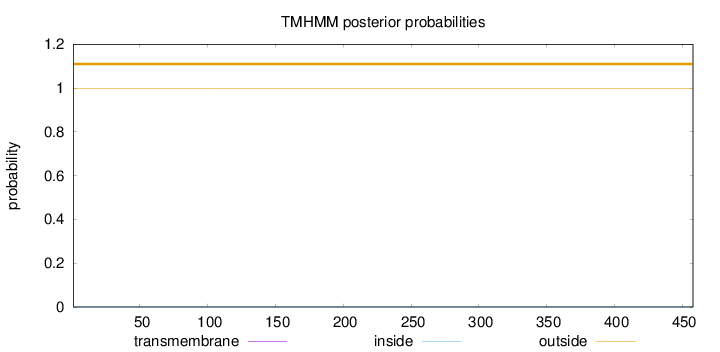
Length:
458
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00151
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00081
outside
1 - 458
Population Genetic Test Statistics
Pi
21.361103
Theta
21.880897
Tajima's D
-0.635001
CLR
0.755647
CSRT
0.212239388030598
Interpretation
Possibly Positive selection
