Gene
KWMTBOMO02312
Annotation
AF461149_1_transposase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.233 Mitochondrial Reliability : 1.366 Nuclear Reliability : 1.379
Sequence
CDS
ATGTTCAATAACAGGCACTGGGTATTCCAACAAGATTCGGCGCCAGCTCATAGAGCGAAGAGCACACAAGACTGGCTGGCGGCGCGTGAAATCGACTTCATTCGGCACGAAGACTGGCCCTCCTCCAGTCCAGATTTGAATCCGTTAGATTACAAGATATGGCAACACTTGGAGGAAAAGGCGTGCTCAAAGCCTCATCCCAATTTGGAGTCACTGAAGACATCCTTGATTAAGGCAGCCGCCGATATTGACATGGACTTCGTTCGTGCTGCGATAGACGACTGGCCGCGCAGATTGAAGGCCTGTATTCAAAATCACGGAGGTCATTTTGAATAA
Protein
MFNNRHWVFQQDSAPAHRAKSTQDWLAAREIDFIRHEDWPSSSPDLNPLDYKIWQHLEEKACSKPHPNLESLKTSLIKAAADIDMDFVRAAIDDWPRRLKACIQNHGGHFE
Summary
Uniprot
Q8ITJ9
B1Q3L9
B9A8E5
B1Q3L5
B1Q3L8
P90693
+ More
A0A226D9B6 A0A1I7RIM5 A0A1I7T5D4 A0A1I7SJQ8 A0A0N4YHH1 A0A0N4XI93 A0A2G5T9N6 E3NDP6 G3G3K1 A0A2H2JHK7 A0A0N4Y729 A0A016S9F8 A0A2G5SZY0 A0A016S0M1 A0A0C2CZV2 A0A0N4XQJ3 E3NJL6 A0A183C618 A0A2G5VIT1 E3M065 E3MBG9 A0A0N4YLD6 A0A0N4XXA5 A0A2G5TSG2 A0A183CKH9 A0A368H7I1 W2STE5 A0A016W3L5 W6NE86 A0A2H2I0K4 H3EUR4 A0A368FVD9 E3MXK7 E3MXK8 E3N9D7 A0A016T5R9 E3M897 E3N1H1 A0A0N4XTP7 A0A1I7RU52 E3MQK2 E3M1J2 E3ND45 E3NHD7 A0A0N4YD61 A0A2G5SF52 A0A0N4XUY5 E3NHK7 A0A016WKB6 E3N6E8 A0A183F5L1 A0A3P7TF39 E3N4H8 A0A183FPD3 A0A3P7ZA30 A0A016TV39 A0A0N4Y0X0 A0A2H2IVX6 A0A183GAB9 A0A3P8EW48 H3ESY7 A0A0N4Y722 H3E4E9 H3EXA9 H3DZR2 H3FB13 H3FES1 A0A2H2J0T6 A0A2G5TZM3 A0A0N4Y9Q8 H3E1D2 B6IKQ6 A0A016TQR4 A0A0C2H248 E3N749 H3DS69 A0A0C2DMD3 A0A0N4YNS9 A0A016WY15 A0A2G5TN79 A0A0C2FMR5 A0A0C2DYF2 A0A016WN34 A0A016VJ32 A0A1I8CC25 A0A016VAC4 H3EE07 A0A2G5TZ20 H3F5A8 A0A183F3N6 A0A3P7TDY7 A0A2G5TUS4
A0A226D9B6 A0A1I7RIM5 A0A1I7T5D4 A0A1I7SJQ8 A0A0N4YHH1 A0A0N4XI93 A0A2G5T9N6 E3NDP6 G3G3K1 A0A2H2JHK7 A0A0N4Y729 A0A016S9F8 A0A2G5SZY0 A0A016S0M1 A0A0C2CZV2 A0A0N4XQJ3 E3NJL6 A0A183C618 A0A2G5VIT1 E3M065 E3MBG9 A0A0N4YLD6 A0A0N4XXA5 A0A2G5TSG2 A0A183CKH9 A0A368H7I1 W2STE5 A0A016W3L5 W6NE86 A0A2H2I0K4 H3EUR4 A0A368FVD9 E3MXK7 E3MXK8 E3N9D7 A0A016T5R9 E3M897 E3N1H1 A0A0N4XTP7 A0A1I7RU52 E3MQK2 E3M1J2 E3ND45 E3NHD7 A0A0N4YD61 A0A2G5SF52 A0A0N4XUY5 E3NHK7 A0A016WKB6 E3N6E8 A0A183F5L1 A0A3P7TF39 E3N4H8 A0A183FPD3 A0A3P7ZA30 A0A016TV39 A0A0N4Y0X0 A0A2H2IVX6 A0A183GAB9 A0A3P8EW48 H3ESY7 A0A0N4Y722 H3E4E9 H3EXA9 H3DZR2 H3FB13 H3FES1 A0A2H2J0T6 A0A2G5TZM3 A0A0N4Y9Q8 H3E1D2 B6IKQ6 A0A016TQR4 A0A0C2H248 E3N749 H3DS69 A0A0C2DMD3 A0A0N4YNS9 A0A016WY15 A0A2G5TN79 A0A0C2FMR5 A0A0C2DYF2 A0A016WN34 A0A016VJ32 A0A1I8CC25 A0A016VAC4 H3EE07 A0A2G5TZ20 H3F5A8 A0A183F3N6 A0A3P7TDY7 A0A2G5TUS4
EMBL
AF461149
AAN06610.1
AB363028
BAG15927.1
AB473770
BAH20555.1
+ More
AB363006 AB363010 AB363014 BAG15923.1 BAG15924.1 AB363018 BAG15926.1 U47917 AAB47739.1 LNIX01000030 OXA41211.1 UYSL01022144 VDL79905.1 UYSL01002425 VDL65835.1 PDUG01000005 PIC23776.1 DS268612 EFO94085.1 JF779677 AEO90418.1 UYSL01020634 VDL75527.1 JARK01001601 EYB87313.1 PDUG01000006 PIC20538.1 JARK01001658 EYB84158.1 KN729202 KIH62598.1 UYSL01009772 VDL68385.1 DS268747 EFP00909.1 PDUG01000001 PIC51673.1 DS268420 EFO87874.1 DS268433 EFO97645.1 UYSL01023059 VDL81638.1 UYSL01019905 VDL71189.1 PIC29926.1 JOJR01000015 RCN51257.1 KI662043 ETN73029.1 JARK01001337 EYC34439.1 CAVP010058755 CDL95145.1 JOJR01000583 RCN36166.1 DS268492 EFP11689.1 EFP11674.1 DS268565 EFO90304.1 JARK01001471 EYB97976.1 DS268428 EFO94379.1 DS268508 EFO83264.1 UYSL01019770 VDL69590.1 DS268466 EFP06959.1 DS268421 EFO88618.1 DS268606 EFO93488.1 DS268673 EFO98001.1 UYSL01021399 VDL78127.1 PDUG01000012 PIC13506.1 UYSL01019810 VDL70150.1 DS268679 EFO98285.1 JARK01000255 EYC39473.1 DS268539 EFO88027.1 UZAH01001595 VDO19836.1 DS268525 EFO85497.1 UZAH01026445 VDO80847.1 JARK01001411 EYC06502.1 UYSL01020109 VDL72812.1 UZAH01031030 VDP13493.1 UYSL01020631 VDL75518.1 PDUG01000004 PIC32571.1 UYSL01020934 VDL76658.1 HE601413 CAS00486.1 JARK01001418 EYC05389.1 KN726559 KIH67820.1 DS268545 EFO88382.1 KN728435 KIH63782.1 UYSL01023743 VDL82626.1 JARK01000059 EYC44525.1 PIC28734.1 KN769991 KIH46151.1 KN726483 KIH68032.1 JARK01000176 EYC41234.1 JARK01001345 EYC27589.1 JARK01001350 EYC23962.1 PIC32570.1 UZAH01000669 VDO19161.1 PIC31069.1
AB363006 AB363010 AB363014 BAG15923.1 BAG15924.1 AB363018 BAG15926.1 U47917 AAB47739.1 LNIX01000030 OXA41211.1 UYSL01022144 VDL79905.1 UYSL01002425 VDL65835.1 PDUG01000005 PIC23776.1 DS268612 EFO94085.1 JF779677 AEO90418.1 UYSL01020634 VDL75527.1 JARK01001601 EYB87313.1 PDUG01000006 PIC20538.1 JARK01001658 EYB84158.1 KN729202 KIH62598.1 UYSL01009772 VDL68385.1 DS268747 EFP00909.1 PDUG01000001 PIC51673.1 DS268420 EFO87874.1 DS268433 EFO97645.1 UYSL01023059 VDL81638.1 UYSL01019905 VDL71189.1 PIC29926.1 JOJR01000015 RCN51257.1 KI662043 ETN73029.1 JARK01001337 EYC34439.1 CAVP010058755 CDL95145.1 JOJR01000583 RCN36166.1 DS268492 EFP11689.1 EFP11674.1 DS268565 EFO90304.1 JARK01001471 EYB97976.1 DS268428 EFO94379.1 DS268508 EFO83264.1 UYSL01019770 VDL69590.1 DS268466 EFP06959.1 DS268421 EFO88618.1 DS268606 EFO93488.1 DS268673 EFO98001.1 UYSL01021399 VDL78127.1 PDUG01000012 PIC13506.1 UYSL01019810 VDL70150.1 DS268679 EFO98285.1 JARK01000255 EYC39473.1 DS268539 EFO88027.1 UZAH01001595 VDO19836.1 DS268525 EFO85497.1 UZAH01026445 VDO80847.1 JARK01001411 EYC06502.1 UYSL01020109 VDL72812.1 UZAH01031030 VDP13493.1 UYSL01020631 VDL75518.1 PDUG01000004 PIC32571.1 UYSL01020934 VDL76658.1 HE601413 CAS00486.1 JARK01001418 EYC05389.1 KN726559 KIH67820.1 DS268545 EFO88382.1 KN728435 KIH63782.1 UYSL01023743 VDL82626.1 JARK01000059 EYC44525.1 PIC28734.1 KN769991 KIH46151.1 KN726483 KIH68032.1 JARK01000176 EYC41234.1 JARK01001345 EYC27589.1 JARK01001350 EYC23962.1 PIC32570.1 UZAH01000669 VDO19161.1 PIC31069.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
Q8ITJ9
B1Q3L9
B9A8E5
B1Q3L5
B1Q3L8
P90693
+ More
A0A226D9B6 A0A1I7RIM5 A0A1I7T5D4 A0A1I7SJQ8 A0A0N4YHH1 A0A0N4XI93 A0A2G5T9N6 E3NDP6 G3G3K1 A0A2H2JHK7 A0A0N4Y729 A0A016S9F8 A0A2G5SZY0 A0A016S0M1 A0A0C2CZV2 A0A0N4XQJ3 E3NJL6 A0A183C618 A0A2G5VIT1 E3M065 E3MBG9 A0A0N4YLD6 A0A0N4XXA5 A0A2G5TSG2 A0A183CKH9 A0A368H7I1 W2STE5 A0A016W3L5 W6NE86 A0A2H2I0K4 H3EUR4 A0A368FVD9 E3MXK7 E3MXK8 E3N9D7 A0A016T5R9 E3M897 E3N1H1 A0A0N4XTP7 A0A1I7RU52 E3MQK2 E3M1J2 E3ND45 E3NHD7 A0A0N4YD61 A0A2G5SF52 A0A0N4XUY5 E3NHK7 A0A016WKB6 E3N6E8 A0A183F5L1 A0A3P7TF39 E3N4H8 A0A183FPD3 A0A3P7ZA30 A0A016TV39 A0A0N4Y0X0 A0A2H2IVX6 A0A183GAB9 A0A3P8EW48 H3ESY7 A0A0N4Y722 H3E4E9 H3EXA9 H3DZR2 H3FB13 H3FES1 A0A2H2J0T6 A0A2G5TZM3 A0A0N4Y9Q8 H3E1D2 B6IKQ6 A0A016TQR4 A0A0C2H248 E3N749 H3DS69 A0A0C2DMD3 A0A0N4YNS9 A0A016WY15 A0A2G5TN79 A0A0C2FMR5 A0A0C2DYF2 A0A016WN34 A0A016VJ32 A0A1I8CC25 A0A016VAC4 H3EE07 A0A2G5TZ20 H3F5A8 A0A183F3N6 A0A3P7TDY7 A0A2G5TUS4
A0A226D9B6 A0A1I7RIM5 A0A1I7T5D4 A0A1I7SJQ8 A0A0N4YHH1 A0A0N4XI93 A0A2G5T9N6 E3NDP6 G3G3K1 A0A2H2JHK7 A0A0N4Y729 A0A016S9F8 A0A2G5SZY0 A0A016S0M1 A0A0C2CZV2 A0A0N4XQJ3 E3NJL6 A0A183C618 A0A2G5VIT1 E3M065 E3MBG9 A0A0N4YLD6 A0A0N4XXA5 A0A2G5TSG2 A0A183CKH9 A0A368H7I1 W2STE5 A0A016W3L5 W6NE86 A0A2H2I0K4 H3EUR4 A0A368FVD9 E3MXK7 E3MXK8 E3N9D7 A0A016T5R9 E3M897 E3N1H1 A0A0N4XTP7 A0A1I7RU52 E3MQK2 E3M1J2 E3ND45 E3NHD7 A0A0N4YD61 A0A2G5SF52 A0A0N4XUY5 E3NHK7 A0A016WKB6 E3N6E8 A0A183F5L1 A0A3P7TF39 E3N4H8 A0A183FPD3 A0A3P7ZA30 A0A016TV39 A0A0N4Y0X0 A0A2H2IVX6 A0A183GAB9 A0A3P8EW48 H3ESY7 A0A0N4Y722 H3E4E9 H3EXA9 H3DZR2 H3FB13 H3FES1 A0A2H2J0T6 A0A2G5TZM3 A0A0N4Y9Q8 H3E1D2 B6IKQ6 A0A016TQR4 A0A0C2H248 E3N749 H3DS69 A0A0C2DMD3 A0A0N4YNS9 A0A016WY15 A0A2G5TN79 A0A0C2FMR5 A0A0C2DYF2 A0A016WN34 A0A016VJ32 A0A1I8CC25 A0A016VAC4 H3EE07 A0A2G5TZ20 H3F5A8 A0A183F3N6 A0A3P7TDY7 A0A2G5TUS4
PDB
5CR4
E-value=4.46823e-09,
Score=139
Ontologies
Topology
Subcellular location
Nucleus
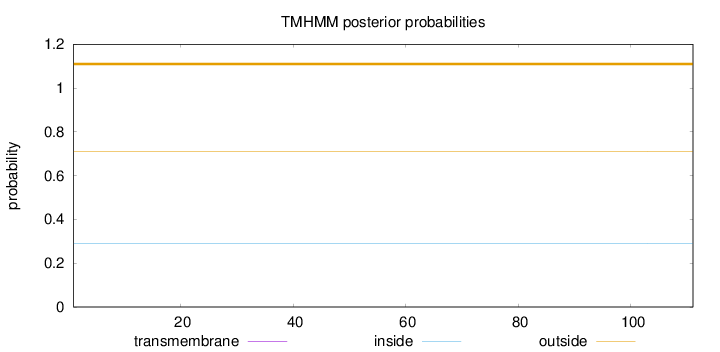
Length:
111
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.29137
outside
1 - 111
Population Genetic Test Statistics
Pi
218.963845
Theta
110.059812
Tajima's D
3.104638
CLR
0
CSRT
0.984050797460127
Interpretation
Uncertain
