Pre Gene Modal
BGIBMGA003785
Annotation
PREDICTED:_mitochondrial_fission_1_protein_[Amyelois_transitella]
Full name
Mitochondrial fission 1 protein
Location in the cell
Cytoplasmic Reliability : 2.126
Sequence
CDS
ATGGAAGACGTATTAGACGAGATTGTTTCATCGGAAGATTTGCAAAAATTCGAGAGGGTCTTTCATGAACAATTGCATCAAGGGAACGTTTCCCACAAAGCCCAGTTTGAATACGCATGGTGTTTAGTGCGCAGCAAGTATCCAACGGATATACGAAAGGGAATCTTACTTCTTAAAGAATTGTTCAACTCTCATCCTGAAGGCAAACGTGACTACCTTTTCTACCTTGCCATTGGTAATGCCAGAATAAAGGAGTACAATAAGGCTTTGCATTATGTGAAATCATTCCTCGAGATTGAACCCGCTAATCAGCAAGTATTGGCATTAGAGCGTCAAATCAATAAACGTATGGAAAAAGAAGGTTTAATTGGAATGGCGGTGGCAGGAGGAGCTGTCCTTGCCCTTGGAGGGCTCGTGGGACTTGGAATCGCACTTGCTTCAAAGAAATAG
Protein
MEDVLDEIVSSEDLQKFERVFHEQLHQGNVSHKAQFEYAWCLVRSKYPTDIRKGILLLKELFNSHPEGKRDYLFYLAIGNARIKEYNKALHYVKSFLEIEPANQQVLALERQINKRMEKEGLIGMAVAGGAVLALGGLVGLGIALASKK
Summary
Description
Involved in the fragmentation of the mitochondrial network and its perinuclear clustering.
Similarity
Belongs to the FIS1 family.
Feature
chain Mitochondrial fission 1 protein
Uniprot
H9J2P8
A0A2A4K8Q6
A0A2H1V9E7
A0A194QQ48
A0A194QGV0
I4DK50
+ More
A0A212EML6 A0A2P8YEY7 A0A0T6B8V0 V5GWU6 A0A1B6EJQ5 A0A195D6N9 F4X6E3 A0A195BCM7 A0A195FNG1 A0A158NJT3 A0A1B6HSA0 A0A151X6B1 A0A1Y1L9B1 E2A8I6 A0A195DX43 D6WNF0 E9IFU4 A0A088A4V0 A0A0L7L0F3 A0A1W4XS76 B1GS86 N6T3Q9 E2BH00 K7IN41 A0A232F9C9 A0A0M9AAZ3 A0A026WR51 A0A1B6EF64 A0A0K8TTR4 V9IGW5 A0A0N7Z9A1 T1HPA9 A0A164Y401 A0A0P4Y632 A0A0P5XFD2 A0A0P5LTN9 A0A224XZA0 A0A0V0G3W9 A0A023FA52 A0A0P6A083 A0A0P5YYZ7 A0A170ZI66 A0A0P6HFD5 A0A0P6D7X8 A0A0P5RXC6 A0A0P5MQP7 A0A0P5L440 A0A0P5QQH5 A0A0P5VYY0 A0A0P6IJF1 A0A2A3EH44 A0A0P4XRB1 A0A0P5TDT1 A0A0N8CB43 A0A1B0CT81 A0A0P5WK00 A0A0P5QF11 A0A0P6BZQ8 E0VVN5 A0A1L8E421 A0A0P6DIX1 A0A0P5AQW5 A0A0P6D7X6 A0A1B0DNL5 R4G3Z5 A0A067R5C8 A0A0P4YRD4 E9HNB5 A0A0P4YSU5 A0A182F2E4 A0A0P5V305 A0A2M3ZHL1 A0A0P5V247 A0A0P5TW51 A0A3Q0JCH8 A0A1S3DDA3 A0A2M4C2H4 A0A023FTU2 A0A0P4YI54 A0A0P5IV43 A0A131YQN8 A0A023GEI4 A0A0C9SCE2 A0A2M4A9J5 A0A224YWI2 L7M6B3 W5JSI9 A0A1E1WXJ6 A0A023FRC4 A0A2M4CY35 A0A0P5QMU1 A0A2M3ZHR4 A0A182SVI2 A0A0P5WZR8
A0A212EML6 A0A2P8YEY7 A0A0T6B8V0 V5GWU6 A0A1B6EJQ5 A0A195D6N9 F4X6E3 A0A195BCM7 A0A195FNG1 A0A158NJT3 A0A1B6HSA0 A0A151X6B1 A0A1Y1L9B1 E2A8I6 A0A195DX43 D6WNF0 E9IFU4 A0A088A4V0 A0A0L7L0F3 A0A1W4XS76 B1GS86 N6T3Q9 E2BH00 K7IN41 A0A232F9C9 A0A0M9AAZ3 A0A026WR51 A0A1B6EF64 A0A0K8TTR4 V9IGW5 A0A0N7Z9A1 T1HPA9 A0A164Y401 A0A0P4Y632 A0A0P5XFD2 A0A0P5LTN9 A0A224XZA0 A0A0V0G3W9 A0A023FA52 A0A0P6A083 A0A0P5YYZ7 A0A170ZI66 A0A0P6HFD5 A0A0P6D7X8 A0A0P5RXC6 A0A0P5MQP7 A0A0P5L440 A0A0P5QQH5 A0A0P5VYY0 A0A0P6IJF1 A0A2A3EH44 A0A0P4XRB1 A0A0P5TDT1 A0A0N8CB43 A0A1B0CT81 A0A0P5WK00 A0A0P5QF11 A0A0P6BZQ8 E0VVN5 A0A1L8E421 A0A0P6DIX1 A0A0P5AQW5 A0A0P6D7X6 A0A1B0DNL5 R4G3Z5 A0A067R5C8 A0A0P4YRD4 E9HNB5 A0A0P4YSU5 A0A182F2E4 A0A0P5V305 A0A2M3ZHL1 A0A0P5V247 A0A0P5TW51 A0A3Q0JCH8 A0A1S3DDA3 A0A2M4C2H4 A0A023FTU2 A0A0P4YI54 A0A0P5IV43 A0A131YQN8 A0A023GEI4 A0A0C9SCE2 A0A2M4A9J5 A0A224YWI2 L7M6B3 W5JSI9 A0A1E1WXJ6 A0A023FRC4 A0A2M4CY35 A0A0P5QMU1 A0A2M3ZHR4 A0A182SVI2 A0A0P5WZR8
Pubmed
EMBL
BABH01034455
NWSH01000050
PCG80193.1
ODYU01001369
SOQ37447.1
KQ461191
+ More
KPJ07090.1 KQ458860 KPJ04768.1 AK401668 BAM18290.1 AGBW02013812 OWR42728.1 PYGN01000651 PSN42819.1 LJIG01009096 KRT83764.1 GALX01002264 JAB66202.1 GECZ01031624 JAS38145.1 KQ976818 KYN08089.1 GL888812 EGI57983.1 KQ976526 KYM81949.1 KQ981424 KYN41847.1 ADTU01018172 GECU01030176 JAS77530.1 KQ982482 KYQ55921.1 GEZM01063919 JAV68910.1 GL437616 EFN70256.1 KQ980155 KYN17453.1 KQ971343 EFA04363.1 GL762903 EFZ20529.1 JTDY01003814 KOB68967.1 AM492666 CAM34497.1 APGK01044799 APGK01044800 APGK01044801 APGK01044802 KB741028 KB632297 ENN74799.1 ERL91708.1 GL448255 EFN85033.1 NNAY01000599 OXU27464.1 KQ435696 KOX80827.1 KK107144 QOIP01000003 EZA57579.1 RLU24355.1 GEDC01000721 JAS36577.1 GDAI01000065 JAI17538.1 JR047173 AEY60343.1 GDKW01001123 JAI55472.1 ACPB03001482 LRGB01000944 KZS14858.1 GDIP01233054 JAI90347.1 GDIP01085220 JAM18495.1 GDIQ01175533 JAK76192.1 GFTR01002504 JAW13922.1 GECL01003368 JAP02756.1 GBBI01000833 JAC17879.1 GDIP01037325 JAM66390.1 GDIP01051276 JAM52439.1 GEMB01002173 JAS01007.1 GDIQ01033389 JAN61348.1 GDIQ01081470 JAN13267.1 GDIQ01100162 JAL51564.1 GDIQ01175532 JAK76193.1 GDIQ01175534 JAK76191.1 GDIQ01110958 JAL40768.1 GDIP01107766 GDIQ01046885 JAL95948.1 JAN47852.1 GDIQ01010079 JAN84658.1 KZ288251 PBC31047.1 GDIP01238754 JAI84647.1 GDIP01127564 JAL76150.1 GDIQ01097115 JAL54611.1 AJWK01027275 AJWK01027276 GDIP01085219 JAM18496.1 GDIQ01136354 JAL15372.1 GDIP01008148 JAM95567.1 DS235812 EEB17441.1 GFDF01000799 JAV13285.1 GDIQ01079871 GDIQ01061979 JAN14866.1 GDIP01195256 JAJ28146.1 GDIQ01081469 JAN13268.1 AJVK01001409 GAHY01001236 JAA76274.1 KK852691 KDR18360.1 GDIP01224899 JAI98502.1 GL732695 EFX66777.1 GDIP01224951 JAI98450.1 GDIP01105165 JAL98549.1 GGFM01007248 MBW27999.1 GDIP01105164 JAL98550.1 GDIP01121112 JAL82602.1 GGFJ01010371 MBW59512.1 GBBL01002223 JAC25097.1 GDIP01227403 JAI95998.1 GDIQ01216125 JAK35600.1 GEDV01006988 JAP81569.1 GBBM01002957 JAC32461.1 GBZX01002437 JAG90303.1 GGFK01004153 MBW37474.1 GFPF01007805 MAA18951.1 GACK01006341 JAA58693.1 ADMH02000292 ETN67096.1 GFAC01007476 JAT91712.1 GBBK01001307 JAC23175.1 GGFL01005600 MBW69778.1 GDIQ01112607 JAL39119.1 GGFM01007300 MBW28051.1 GDIP01079267 JAM24448.1
KPJ07090.1 KQ458860 KPJ04768.1 AK401668 BAM18290.1 AGBW02013812 OWR42728.1 PYGN01000651 PSN42819.1 LJIG01009096 KRT83764.1 GALX01002264 JAB66202.1 GECZ01031624 JAS38145.1 KQ976818 KYN08089.1 GL888812 EGI57983.1 KQ976526 KYM81949.1 KQ981424 KYN41847.1 ADTU01018172 GECU01030176 JAS77530.1 KQ982482 KYQ55921.1 GEZM01063919 JAV68910.1 GL437616 EFN70256.1 KQ980155 KYN17453.1 KQ971343 EFA04363.1 GL762903 EFZ20529.1 JTDY01003814 KOB68967.1 AM492666 CAM34497.1 APGK01044799 APGK01044800 APGK01044801 APGK01044802 KB741028 KB632297 ENN74799.1 ERL91708.1 GL448255 EFN85033.1 NNAY01000599 OXU27464.1 KQ435696 KOX80827.1 KK107144 QOIP01000003 EZA57579.1 RLU24355.1 GEDC01000721 JAS36577.1 GDAI01000065 JAI17538.1 JR047173 AEY60343.1 GDKW01001123 JAI55472.1 ACPB03001482 LRGB01000944 KZS14858.1 GDIP01233054 JAI90347.1 GDIP01085220 JAM18495.1 GDIQ01175533 JAK76192.1 GFTR01002504 JAW13922.1 GECL01003368 JAP02756.1 GBBI01000833 JAC17879.1 GDIP01037325 JAM66390.1 GDIP01051276 JAM52439.1 GEMB01002173 JAS01007.1 GDIQ01033389 JAN61348.1 GDIQ01081470 JAN13267.1 GDIQ01100162 JAL51564.1 GDIQ01175532 JAK76193.1 GDIQ01175534 JAK76191.1 GDIQ01110958 JAL40768.1 GDIP01107766 GDIQ01046885 JAL95948.1 JAN47852.1 GDIQ01010079 JAN84658.1 KZ288251 PBC31047.1 GDIP01238754 JAI84647.1 GDIP01127564 JAL76150.1 GDIQ01097115 JAL54611.1 AJWK01027275 AJWK01027276 GDIP01085219 JAM18496.1 GDIQ01136354 JAL15372.1 GDIP01008148 JAM95567.1 DS235812 EEB17441.1 GFDF01000799 JAV13285.1 GDIQ01079871 GDIQ01061979 JAN14866.1 GDIP01195256 JAJ28146.1 GDIQ01081469 JAN13268.1 AJVK01001409 GAHY01001236 JAA76274.1 KK852691 KDR18360.1 GDIP01224899 JAI98502.1 GL732695 EFX66777.1 GDIP01224951 JAI98450.1 GDIP01105165 JAL98549.1 GGFM01007248 MBW27999.1 GDIP01105164 JAL98550.1 GDIP01121112 JAL82602.1 GGFJ01010371 MBW59512.1 GBBL01002223 JAC25097.1 GDIP01227403 JAI95998.1 GDIQ01216125 JAK35600.1 GEDV01006988 JAP81569.1 GBBM01002957 JAC32461.1 GBZX01002437 JAG90303.1 GGFK01004153 MBW37474.1 GFPF01007805 MAA18951.1 GACK01006341 JAA58693.1 ADMH02000292 ETN67096.1 GFAC01007476 JAT91712.1 GBBK01001307 JAC23175.1 GGFL01005600 MBW69778.1 GDIQ01112607 JAL39119.1 GGFM01007300 MBW28051.1 GDIP01079267 JAM24448.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000245037
+ More
UP000078542 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000000311 UP000078492 UP000007266 UP000005203 UP000037510 UP000192223 UP000019118 UP000030742 UP000008237 UP000002358 UP000215335 UP000053105 UP000053097 UP000279307 UP000015103 UP000076858 UP000242457 UP000092461 UP000009046 UP000092462 UP000027135 UP000000305 UP000069272 UP000079169 UP000000673 UP000075901
UP000078542 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000000311 UP000078492 UP000007266 UP000005203 UP000037510 UP000192223 UP000019118 UP000030742 UP000008237 UP000002358 UP000215335 UP000053105 UP000053097 UP000279307 UP000015103 UP000076858 UP000242457 UP000092461 UP000009046 UP000092462 UP000027135 UP000000305 UP000069272 UP000079169 UP000000673 UP000075901
PRIDE
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
CDD
ProteinModelPortal
H9J2P8
A0A2A4K8Q6
A0A2H1V9E7
A0A194QQ48
A0A194QGV0
I4DK50
+ More
A0A212EML6 A0A2P8YEY7 A0A0T6B8V0 V5GWU6 A0A1B6EJQ5 A0A195D6N9 F4X6E3 A0A195BCM7 A0A195FNG1 A0A158NJT3 A0A1B6HSA0 A0A151X6B1 A0A1Y1L9B1 E2A8I6 A0A195DX43 D6WNF0 E9IFU4 A0A088A4V0 A0A0L7L0F3 A0A1W4XS76 B1GS86 N6T3Q9 E2BH00 K7IN41 A0A232F9C9 A0A0M9AAZ3 A0A026WR51 A0A1B6EF64 A0A0K8TTR4 V9IGW5 A0A0N7Z9A1 T1HPA9 A0A164Y401 A0A0P4Y632 A0A0P5XFD2 A0A0P5LTN9 A0A224XZA0 A0A0V0G3W9 A0A023FA52 A0A0P6A083 A0A0P5YYZ7 A0A170ZI66 A0A0P6HFD5 A0A0P6D7X8 A0A0P5RXC6 A0A0P5MQP7 A0A0P5L440 A0A0P5QQH5 A0A0P5VYY0 A0A0P6IJF1 A0A2A3EH44 A0A0P4XRB1 A0A0P5TDT1 A0A0N8CB43 A0A1B0CT81 A0A0P5WK00 A0A0P5QF11 A0A0P6BZQ8 E0VVN5 A0A1L8E421 A0A0P6DIX1 A0A0P5AQW5 A0A0P6D7X6 A0A1B0DNL5 R4G3Z5 A0A067R5C8 A0A0P4YRD4 E9HNB5 A0A0P4YSU5 A0A182F2E4 A0A0P5V305 A0A2M3ZHL1 A0A0P5V247 A0A0P5TW51 A0A3Q0JCH8 A0A1S3DDA3 A0A2M4C2H4 A0A023FTU2 A0A0P4YI54 A0A0P5IV43 A0A131YQN8 A0A023GEI4 A0A0C9SCE2 A0A2M4A9J5 A0A224YWI2 L7M6B3 W5JSI9 A0A1E1WXJ6 A0A023FRC4 A0A2M4CY35 A0A0P5QMU1 A0A2M3ZHR4 A0A182SVI2 A0A0P5WZR8
A0A212EML6 A0A2P8YEY7 A0A0T6B8V0 V5GWU6 A0A1B6EJQ5 A0A195D6N9 F4X6E3 A0A195BCM7 A0A195FNG1 A0A158NJT3 A0A1B6HSA0 A0A151X6B1 A0A1Y1L9B1 E2A8I6 A0A195DX43 D6WNF0 E9IFU4 A0A088A4V0 A0A0L7L0F3 A0A1W4XS76 B1GS86 N6T3Q9 E2BH00 K7IN41 A0A232F9C9 A0A0M9AAZ3 A0A026WR51 A0A1B6EF64 A0A0K8TTR4 V9IGW5 A0A0N7Z9A1 T1HPA9 A0A164Y401 A0A0P4Y632 A0A0P5XFD2 A0A0P5LTN9 A0A224XZA0 A0A0V0G3W9 A0A023FA52 A0A0P6A083 A0A0P5YYZ7 A0A170ZI66 A0A0P6HFD5 A0A0P6D7X8 A0A0P5RXC6 A0A0P5MQP7 A0A0P5L440 A0A0P5QQH5 A0A0P5VYY0 A0A0P6IJF1 A0A2A3EH44 A0A0P4XRB1 A0A0P5TDT1 A0A0N8CB43 A0A1B0CT81 A0A0P5WK00 A0A0P5QF11 A0A0P6BZQ8 E0VVN5 A0A1L8E421 A0A0P6DIX1 A0A0P5AQW5 A0A0P6D7X6 A0A1B0DNL5 R4G3Z5 A0A067R5C8 A0A0P4YRD4 E9HNB5 A0A0P4YSU5 A0A182F2E4 A0A0P5V305 A0A2M3ZHL1 A0A0P5V247 A0A0P5TW51 A0A3Q0JCH8 A0A1S3DDA3 A0A2M4C2H4 A0A023FTU2 A0A0P4YI54 A0A0P5IV43 A0A131YQN8 A0A023GEI4 A0A0C9SCE2 A0A2M4A9J5 A0A224YWI2 L7M6B3 W5JSI9 A0A1E1WXJ6 A0A023FRC4 A0A2M4CY35 A0A0P5QMU1 A0A2M3ZHR4 A0A182SVI2 A0A0P5WZR8
PDB
1PC2
E-value=2.55726e-31,
Score=332
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion outer membrane
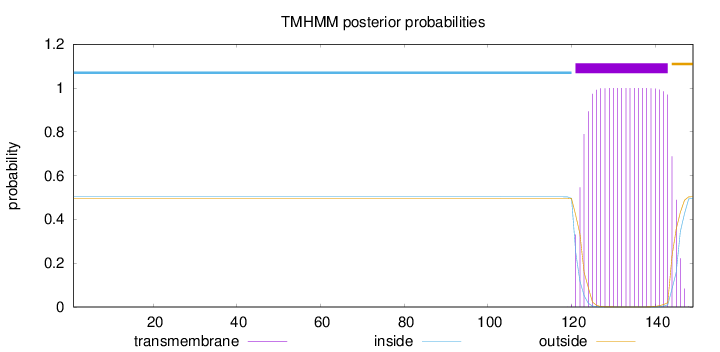
Length:
149
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.96095
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.50328
inside
1 - 120
TMhelix
121 - 143
outside
144 - 149
Population Genetic Test Statistics
Pi
193.682569
Theta
177.341181
Tajima's D
0.43755
CLR
0
CSRT
0.495575221238938
Interpretation
Uncertain
