Gene
KWMTBOMO02308 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003767
Annotation
autophagy_related_protein_Atg3-like_protein_[Bombyx_mori]
Full name
Autophagy-related protein 3
Location in the cell
Cytoplasmic Reliability : 1.448 Nuclear Reliability : 1.423
Sequence
CDS
ATGCAAAGTGTAATAAACACAGTAAAGGGAACTGCTCTTGGCGTGGCCGGTTACTTGACACCGGTGTTGAAGGAATCGAAATTTCAAGAAACTGGTGTACTGACTCCTGAAGAGTTTGTAGCTGCTGGTGACCACCTGGTACACCACTGCCCTACCTGGCAGTGGGCAAAGGGAGAGGAAGCTAAGATTAGGCCTTACCTTCCTGCAGACAAACAATTCCTTATAACTCGAAATGTGCCATGCTACAGACGATGTAAACAAATTGAATACTGTGAAGATAAAGAGAAAGTGATAGAAGATGAAAACGATGCAGATGGTGGCTGGGTTGATACTCATCACTATGACAATGCAGGCTCCCCTGCACTCGAGGAAAAGGTCTGTGAGATGACCCTCGAAGCAGCTGACACTGGCGACAGTGACGATGGAGCTGCCGACGGTGGTGATGATGATGAAGACGAAGATGATGAGGGCGAGGCTGAGGACATGGAGCAGTTCCAAGAATCTGGTCTTTTAGATGAAGTAGATCCGTCAACAGATCTAGCCCAGCGTACAGAGCTAAAACTCAAAGCCGATAAGAAACAGAAGCAAGGAGAAGGTGATGAGATCGTCCGGACTCGAACTTACGACCTACACATCACTTATGACAAGTATTACCAAACTCCGAGGCTGTGGCTCATCGGATATGATGAGAACCGGTCACTCCTGACTGTGGAACAGATGTACGAAGACGTGTCCCAAGATCACGCTAAAAAGACTGTCACGATGGAATCTCACCCTCATCTCTCTGGACCCAGCATGGCTTCTGTACACCCTTGCAGGCACGCGGACGTCATGAAGAAGATAATAGAGACGGTAACGGAGAGCGGCGGCGAGATGGGCGTCCACTCCTACCTCATAGTATTCCTGAAATTCGTACAAACAGTTATACCCACCATCGAATATGATTTCACACAGAATTTCTCGATTAATTAA
Protein
MQSVINTVKGTALGVAGYLTPVLKESKFQETGVLTPEEFVAAGDHLVHHCPTWQWAKGEEAKIRPYLPADKQFLITRNVPCYRRCKQIEYCEDKEKVIEDENDADGGWVDTHHYDNAGSPALEEKVCEMTLEAADTGDSDDGAADGGDDDEDEDDEGEAEDMEQFQESGLLDEVDPSTDLAQRTELKLKADKKQKQGEGDEIVRTRTYDLHITYDKYYQTPRLWLIGYDENRSLLTVEQMYEDVSQDHAKKTVTMESHPHLSGPSMASVHPCRHADVMKKIIETVTESGGEMGVHSYLIVFLKFVQTVIPTIEYDFTQNFSIN
Summary
Similarity
Belongs to the ATG3 family.
Belongs to the tetraspanin (TM4SF) family.
Belongs to the tetraspanin (TM4SF) family.
Uniprot
H9J2N0
B7TYK4
J9XR17
A0A2A4K951
A0A2H1V9I0
A0A194QU71
+ More
A0A212EMM9 A0A2M4CSF1 A0A182Y619 A0A182X7R3 A0A2M4AQ44 A0A182TR94 Q7QGK7 A0A2M3ZAS5 A0A182VER9 A0A2M4CSK4 A0A182WER1 A0A182QZU0 A0A182JYG5 A0A023EPU3 A0A2M4BTJ9 A0A182RAP8 Q17MR1 A0A182LS77 A0A0K8TSN1 A0A182G9L0 A0A194QGY2 A0A0L7L0E2 U5EY99 B4GRS6 A0A1B6GE40 Q2M0D4 A0A0M4EMQ8 A0A1D2NJG1 B3M873 A0A067QRY9 B0W2I5 A0A182MXQ4 B4KXS6 Q9VVS6 B4PIZ0 D6WMT7 A0A3B0J7P0 B4IFV5 B4QPW0 A0A1L8DW77 D3TMV4 A0A1Q3F8S2 B3NHP1 B4LFX5 A0A182FBI5 B4J1T7 K7IXC6 A0A1B6IFG1 D4AH84 A0A1I8PFE8 A0A0A7A878 A0A1B6KRR4 A0A1W4UBW6 W8C4H3 A0A1Y1LY63 A0A2J7R669 L7M6A5 E2B633 B4N4E9 A0A0K8ULX9 A0A3B3WG89 A0A087YLZ9 A0A0K8SGM0 A0A146LC15 A0A1B6D7P0 A0A0P4VKE6 A0A3P9PWC4 A0A224XNE5 A0A3B3UQ55 A0A0F7LE61 W5M0T0 A0A131XTX4 A0A069DS52 A0A3P8SGH9 A0A3B5AD71 A0A088ATD1 A0A131XEW6 A0A2I4D689 A0A182HN76 A0A147BEM9 A0A2I4D6B1 A0A3B4VKZ9 A0A224YS46 A0A1B6DRF4 A0A1A8SM27 M4A5U4 A0A3P8VA53 A0A3B4Y7K3 B7PSN5 A0A131Z2N0 A0A034WJI0 E2AIM5 A0A1A7YSS7 A0A293MWQ9
A0A212EMM9 A0A2M4CSF1 A0A182Y619 A0A182X7R3 A0A2M4AQ44 A0A182TR94 Q7QGK7 A0A2M3ZAS5 A0A182VER9 A0A2M4CSK4 A0A182WER1 A0A182QZU0 A0A182JYG5 A0A023EPU3 A0A2M4BTJ9 A0A182RAP8 Q17MR1 A0A182LS77 A0A0K8TSN1 A0A182G9L0 A0A194QGY2 A0A0L7L0E2 U5EY99 B4GRS6 A0A1B6GE40 Q2M0D4 A0A0M4EMQ8 A0A1D2NJG1 B3M873 A0A067QRY9 B0W2I5 A0A182MXQ4 B4KXS6 Q9VVS6 B4PIZ0 D6WMT7 A0A3B0J7P0 B4IFV5 B4QPW0 A0A1L8DW77 D3TMV4 A0A1Q3F8S2 B3NHP1 B4LFX5 A0A182FBI5 B4J1T7 K7IXC6 A0A1B6IFG1 D4AH84 A0A1I8PFE8 A0A0A7A878 A0A1B6KRR4 A0A1W4UBW6 W8C4H3 A0A1Y1LY63 A0A2J7R669 L7M6A5 E2B633 B4N4E9 A0A0K8ULX9 A0A3B3WG89 A0A087YLZ9 A0A0K8SGM0 A0A146LC15 A0A1B6D7P0 A0A0P4VKE6 A0A3P9PWC4 A0A224XNE5 A0A3B3UQ55 A0A0F7LE61 W5M0T0 A0A131XTX4 A0A069DS52 A0A3P8SGH9 A0A3B5AD71 A0A088ATD1 A0A131XEW6 A0A2I4D689 A0A182HN76 A0A147BEM9 A0A2I4D6B1 A0A3B4VKZ9 A0A224YS46 A0A1B6DRF4 A0A1A8SM27 M4A5U4 A0A3P8VA53 A0A3B4Y7K3 B7PSN5 A0A131Z2N0 A0A034WJI0 E2AIM5 A0A1A7YSS7 A0A293MWQ9
Pubmed
19121390
19470186
26354079
22118469
25244985
12364791
+ More
14747013 17210077 24945155 17510324 26369729 26483478 26227816 17994087 15632085 27289101 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18362917 19820115 22936249 20353571 20075255 24495485 28004739 25576852 20798317 26823975 27129103 26334808 28049606 29652888 28797301 23542700 24487278 26830274 25348373
14747013 17210077 24945155 17510324 26369729 26483478 26227816 17994087 15632085 27289101 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18362917 19820115 22936249 20353571 20075255 24495485 28004739 25576852 20798317 26823975 27129103 26334808 28049606 29652888 28797301 23542700 24487278 26830274 25348373
EMBL
BABH01034455
FJ416327
ACJ46061.1
JX313129
AFS31123.1
NWSH01000050
+ More
PCG80192.1 ODYU01001369 SOQ37446.1 KQ461191 KPJ07091.1 AGBW02013812 OWR42727.1 GGFL01004082 MBW68260.1 GGFK01009576 MBW42897.1 AAAB01008834 EAA05703.3 GGFM01004883 MBW25634.1 GGFL01004081 MBW68259.1 AXCN02001048 GAPW01002530 JAC11068.1 GGFJ01007192 MBW56333.1 CH477204 EAT47961.1 AXCM01003183 GDAI01000472 JAI17131.1 JXUM01049371 JXUM01049372 KQ561601 KXJ78032.1 KQ458860 KPJ04767.1 JTDY01003814 KOB68968.1 GANO01000424 JAB59447.1 CH479188 EDW40461.1 GECZ01009206 JAS60563.1 CH379069 EAL31000.1 CP012525 ALC44423.1 ALC45035.1 LJIJ01000023 ODN05418.1 CH902618 EDV41012.1 KK853004 KDR12605.1 DS231827 EDS29293.1 CH933809 EDW19783.1 AE014296 AY119557 AAF49233.1 AAM50211.1 CM000159 EDW94581.1 KQ971342 EFA03061.1 OUUW01000002 SPP76183.1 CH480834 EDW46542.1 CM000363 CM002912 EDX10988.1 KMZ00442.1 GFDF01003383 JAV10701.1 EZ422756 ADD19032.1 GFDL01011099 JAV23946.1 CH954178 EDV51836.1 CH940647 EDW70374.1 CH916366 EDV98017.1 AAZX01000032 GECU01032226 GECU01028077 GECU01027654 GECU01022063 GECU01019670 GECU01013822 GECU01000903 JAS75480.1 JAS79629.1 JAS80052.1 JAS85643.1 JAS88036.1 JAS93884.1 JAT06804.1 AB513349 BAI82575.1 KF670693 AHC00664.1 GEBQ01025835 JAT14142.1 GAMC01002308 GAMC01002307 JAC04249.1 GEZM01046348 GEZM01046347 JAV77310.1 NEVH01006980 PNF36327.1 GACK01005474 JAA59560.1 GL445914 EFN88822.1 CH964095 EDW79023.2 GDHF01024753 JAI27561.1 AYCK01022864 GBRD01013896 JAG51930.1 GDHC01020056 GDHC01014519 JAP98572.1 JAQ04110.1 GEDC01026203 GEDC01015591 JAS11095.1 JAS21707.1 GDKW01001050 JAI55545.1 GFTR01005088 JAW11338.1 KP317124 AKH60756.1 AHAT01025099 GEFM01006775 JAP69021.1 GBGD01002119 JAC86770.1 GEFH01004370 JAP64211.1 GEGO01006157 JAR89247.1 GFPF01006275 MAA17421.1 GEDC01020505 GEDC01009047 JAS16793.1 JAS28251.1 HAEI01016489 SBS18958.1 ABJB010403108 ABJB010476986 ABJB010679057 ABJB011130049 DS779597 EEC09607.1 GEDV01003841 JAP84716.1 GAKP01004677 JAC54275.1 GL439840 EFN66714.1 HADW01009460 HADX01010987 SBP33219.1 GFWV01019630 MAA44358.1
PCG80192.1 ODYU01001369 SOQ37446.1 KQ461191 KPJ07091.1 AGBW02013812 OWR42727.1 GGFL01004082 MBW68260.1 GGFK01009576 MBW42897.1 AAAB01008834 EAA05703.3 GGFM01004883 MBW25634.1 GGFL01004081 MBW68259.1 AXCN02001048 GAPW01002530 JAC11068.1 GGFJ01007192 MBW56333.1 CH477204 EAT47961.1 AXCM01003183 GDAI01000472 JAI17131.1 JXUM01049371 JXUM01049372 KQ561601 KXJ78032.1 KQ458860 KPJ04767.1 JTDY01003814 KOB68968.1 GANO01000424 JAB59447.1 CH479188 EDW40461.1 GECZ01009206 JAS60563.1 CH379069 EAL31000.1 CP012525 ALC44423.1 ALC45035.1 LJIJ01000023 ODN05418.1 CH902618 EDV41012.1 KK853004 KDR12605.1 DS231827 EDS29293.1 CH933809 EDW19783.1 AE014296 AY119557 AAF49233.1 AAM50211.1 CM000159 EDW94581.1 KQ971342 EFA03061.1 OUUW01000002 SPP76183.1 CH480834 EDW46542.1 CM000363 CM002912 EDX10988.1 KMZ00442.1 GFDF01003383 JAV10701.1 EZ422756 ADD19032.1 GFDL01011099 JAV23946.1 CH954178 EDV51836.1 CH940647 EDW70374.1 CH916366 EDV98017.1 AAZX01000032 GECU01032226 GECU01028077 GECU01027654 GECU01022063 GECU01019670 GECU01013822 GECU01000903 JAS75480.1 JAS79629.1 JAS80052.1 JAS85643.1 JAS88036.1 JAS93884.1 JAT06804.1 AB513349 BAI82575.1 KF670693 AHC00664.1 GEBQ01025835 JAT14142.1 GAMC01002308 GAMC01002307 JAC04249.1 GEZM01046348 GEZM01046347 JAV77310.1 NEVH01006980 PNF36327.1 GACK01005474 JAA59560.1 GL445914 EFN88822.1 CH964095 EDW79023.2 GDHF01024753 JAI27561.1 AYCK01022864 GBRD01013896 JAG51930.1 GDHC01020056 GDHC01014519 JAP98572.1 JAQ04110.1 GEDC01026203 GEDC01015591 JAS11095.1 JAS21707.1 GDKW01001050 JAI55545.1 GFTR01005088 JAW11338.1 KP317124 AKH60756.1 AHAT01025099 GEFM01006775 JAP69021.1 GBGD01002119 JAC86770.1 GEFH01004370 JAP64211.1 GEGO01006157 JAR89247.1 GFPF01006275 MAA17421.1 GEDC01020505 GEDC01009047 JAS16793.1 JAS28251.1 HAEI01016489 SBS18958.1 ABJB010403108 ABJB010476986 ABJB010679057 ABJB011130049 DS779597 EEC09607.1 GEDV01003841 JAP84716.1 GAKP01004677 JAC54275.1 GL439840 EFN66714.1 HADW01009460 HADX01010987 SBP33219.1 GFWV01019630 MAA44358.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000076408
UP000076407
+ More
UP000075902 UP000007062 UP000075903 UP000075920 UP000075886 UP000075881 UP000075900 UP000008820 UP000075883 UP000069940 UP000249989 UP000053268 UP000037510 UP000008744 UP000001819 UP000092553 UP000094527 UP000007801 UP000027135 UP000002320 UP000075884 UP000009192 UP000000803 UP000002282 UP000007266 UP000268350 UP000001292 UP000000304 UP000008711 UP000008792 UP000069272 UP000001070 UP000002358 UP000095300 UP000192221 UP000235965 UP000008237 UP000007798 UP000261480 UP000028760 UP000242638 UP000261500 UP000018468 UP000265080 UP000261400 UP000005203 UP000192220 UP000261420 UP000002852 UP000265120 UP000261360 UP000001555 UP000000311
UP000075902 UP000007062 UP000075903 UP000075920 UP000075886 UP000075881 UP000075900 UP000008820 UP000075883 UP000069940 UP000249989 UP000053268 UP000037510 UP000008744 UP000001819 UP000092553 UP000094527 UP000007801 UP000027135 UP000002320 UP000075884 UP000009192 UP000000803 UP000002282 UP000007266 UP000268350 UP000001292 UP000000304 UP000008711 UP000008792 UP000069272 UP000001070 UP000002358 UP000095300 UP000192221 UP000235965 UP000008237 UP000007798 UP000261480 UP000028760 UP000242638 UP000261500 UP000018468 UP000265080 UP000261400 UP000005203 UP000192220 UP000261420 UP000002852 UP000265120 UP000261360 UP000001555 UP000000311
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9J2N0
B7TYK4
J9XR17
A0A2A4K951
A0A2H1V9I0
A0A194QU71
+ More
A0A212EMM9 A0A2M4CSF1 A0A182Y619 A0A182X7R3 A0A2M4AQ44 A0A182TR94 Q7QGK7 A0A2M3ZAS5 A0A182VER9 A0A2M4CSK4 A0A182WER1 A0A182QZU0 A0A182JYG5 A0A023EPU3 A0A2M4BTJ9 A0A182RAP8 Q17MR1 A0A182LS77 A0A0K8TSN1 A0A182G9L0 A0A194QGY2 A0A0L7L0E2 U5EY99 B4GRS6 A0A1B6GE40 Q2M0D4 A0A0M4EMQ8 A0A1D2NJG1 B3M873 A0A067QRY9 B0W2I5 A0A182MXQ4 B4KXS6 Q9VVS6 B4PIZ0 D6WMT7 A0A3B0J7P0 B4IFV5 B4QPW0 A0A1L8DW77 D3TMV4 A0A1Q3F8S2 B3NHP1 B4LFX5 A0A182FBI5 B4J1T7 K7IXC6 A0A1B6IFG1 D4AH84 A0A1I8PFE8 A0A0A7A878 A0A1B6KRR4 A0A1W4UBW6 W8C4H3 A0A1Y1LY63 A0A2J7R669 L7M6A5 E2B633 B4N4E9 A0A0K8ULX9 A0A3B3WG89 A0A087YLZ9 A0A0K8SGM0 A0A146LC15 A0A1B6D7P0 A0A0P4VKE6 A0A3P9PWC4 A0A224XNE5 A0A3B3UQ55 A0A0F7LE61 W5M0T0 A0A131XTX4 A0A069DS52 A0A3P8SGH9 A0A3B5AD71 A0A088ATD1 A0A131XEW6 A0A2I4D689 A0A182HN76 A0A147BEM9 A0A2I4D6B1 A0A3B4VKZ9 A0A224YS46 A0A1B6DRF4 A0A1A8SM27 M4A5U4 A0A3P8VA53 A0A3B4Y7K3 B7PSN5 A0A131Z2N0 A0A034WJI0 E2AIM5 A0A1A7YSS7 A0A293MWQ9
A0A212EMM9 A0A2M4CSF1 A0A182Y619 A0A182X7R3 A0A2M4AQ44 A0A182TR94 Q7QGK7 A0A2M3ZAS5 A0A182VER9 A0A2M4CSK4 A0A182WER1 A0A182QZU0 A0A182JYG5 A0A023EPU3 A0A2M4BTJ9 A0A182RAP8 Q17MR1 A0A182LS77 A0A0K8TSN1 A0A182G9L0 A0A194QGY2 A0A0L7L0E2 U5EY99 B4GRS6 A0A1B6GE40 Q2M0D4 A0A0M4EMQ8 A0A1D2NJG1 B3M873 A0A067QRY9 B0W2I5 A0A182MXQ4 B4KXS6 Q9VVS6 B4PIZ0 D6WMT7 A0A3B0J7P0 B4IFV5 B4QPW0 A0A1L8DW77 D3TMV4 A0A1Q3F8S2 B3NHP1 B4LFX5 A0A182FBI5 B4J1T7 K7IXC6 A0A1B6IFG1 D4AH84 A0A1I8PFE8 A0A0A7A878 A0A1B6KRR4 A0A1W4UBW6 W8C4H3 A0A1Y1LY63 A0A2J7R669 L7M6A5 E2B633 B4N4E9 A0A0K8ULX9 A0A3B3WG89 A0A087YLZ9 A0A0K8SGM0 A0A146LC15 A0A1B6D7P0 A0A0P4VKE6 A0A3P9PWC4 A0A224XNE5 A0A3B3UQ55 A0A0F7LE61 W5M0T0 A0A131XTX4 A0A069DS52 A0A3P8SGH9 A0A3B5AD71 A0A088ATD1 A0A131XEW6 A0A2I4D689 A0A182HN76 A0A147BEM9 A0A2I4D6B1 A0A3B4VKZ9 A0A224YS46 A0A1B6DRF4 A0A1A8SM27 M4A5U4 A0A3P8VA53 A0A3B4Y7K3 B7PSN5 A0A131Z2N0 A0A034WJI0 E2AIM5 A0A1A7YSS7 A0A293MWQ9
PDB
3VX8
E-value=9.99129e-65,
Score=625
Ontologies
PATHWAY
GO
Topology
Subcellular location
Cytoplasm
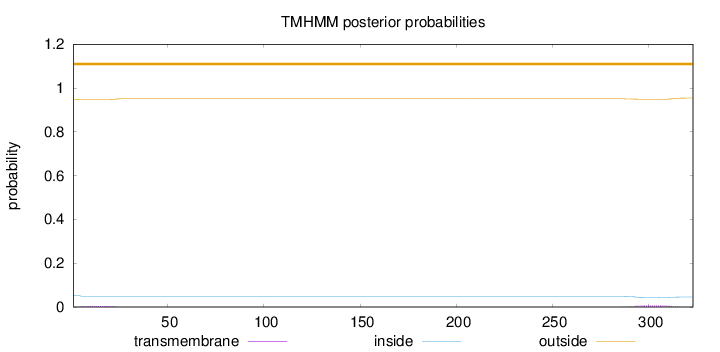
Length:
323
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24131
Exp number, first 60 AAs:
0.08039
Total prob of N-in:
0.05290
outside
1 - 323
Population Genetic Test Statistics
Pi
310.05361
Theta
207.946911
Tajima's D
1.575138
CLR
0
CSRT
0.806859657017149
Interpretation
Uncertain
