Gene
KWMTBOMO02302
Pre Gene Modal
BGIBMGA003771
Annotation
PREDICTED:_group_XV_phospholipase_A2-like_isoform_X1_[Amyelois_transitella]
Location in the cell
Lysosomal Reliability : 1.306 PlasmaMembrane Reliability : 1.123
Sequence
CDS
ATGTTTGGTATTAGAGTAACTTTTCAGTTAATATTTGTTTATTATTTACTGGCCAAGAGCTCTTGGGGACTGTCTCCTGTAATACTTATTCCTGGTGATGGAGGTAATCAATTAGAAGCAAGACTTAACAGATCGTTAGTGGTCCACTACATTTGTAGTAAAACCTCAGCCGACTACTTTAATATATGGTTGAACCTTGAACTTCTCGTGCCTTATATTATTGACTGTTGGGTTGATAATCTAAGATTGGAATATGACAATGTTACGAGAACAACTCACAGCCCCGTTGGAGTTGAAGTCCGAGTTCCAGGCTGGGGTAACACAGAACCAGTTGAATGGTTGGATCCGTCTCATCAGTCATCTGGAGCATATTTCAATAGTATAGCAGATTCTCTAGTCAGCATTGGTTATGTTAGAAATGTATCCATCATAGGAGCTCCGTATGACTTCAGAAAAGCTCCAAATGAAAACCAAGAATTCTTCATAAAGTTTAAAAGCTTAGTAGAAGACACTTATAACAAGAACAATAAGACATCAGTTACCTTAATAGTGCATAGTATGGGAGGCAGAATGGCATTACATTTCCTTCAGCTCCAAACTCAAAAATGGAAGGATCAGTACATAAGAAGGATGATTTCTCTTTCCACACCCTGGGCAGGGTCCATGAAGGCTGTTAAAGTATTTGCTATTGGCGATGATCTCGGCTCAAGGATGTTGTTCGAGAGCACAATGAGGACCCAGCAGATAACCTGTCCCTCGCTGGCCTGGCTGCTACCCTCACCCTTGTTCTGGAAGCCTAATGAAGTTCTGGTGCAAACTGACAAATATAATTACACGATAAACGACTTTCAAAAGTTGTTCAATGACATGGAGCTACCCAACGCTTGGGAGATGAAAAAGGACACTGAGAAATACACAAAGGACTTCAGCGCTCCCGGCGTCGAACTGCACTGCGTGTACGGGTACAACGTCGCCACTGTGGAGCGGTTGGTATACAAGGCTGGCACGTGGCTGGACGGCTACCCCGCGCTGGGCACGGGCGACGGCGACGGCACCGTCAACCTGCGGTCCCTGGGCGCCTGCGAGCACTGGCGGAAACGCCGCGGACGCACCACGCTCAACGGAGGCAAGCCCGTCAAGTCACTGCCACTCAGCAACGCAGAACACCTCAAGATCCTCCACGACAAACGCGTCATCGACTACATCACCACTGTCATGAGCAACTAG
Protein
MFGIRVTFQLIFVYYLLAKSSWGLSPVILIPGDGGNQLEARLNRSLVVHYICSKTSADYFNIWLNLELLVPYIIDCWVDNLRLEYDNVTRTTHSPVGVEVRVPGWGNTEPVEWLDPSHQSSGAYFNSIADSLVSIGYVRNVSIIGAPYDFRKAPNENQEFFIKFKSLVEDTYNKNNKTSVTLIVHSMGGRMALHFLQLQTQKWKDQYIRRMISLSTPWAGSMKAVKVFAIGDDLGSRMLFESTMRTQQITCPSLAWLLPSPLFWKPNEVLVQTDKYNYTINDFQKLFNDMELPNAWEMKKDTEKYTKDFSAPGVELHCVYGYNVATVERLVYKAGTWLDGYPALGTGDGDGTVNLRSLGACEHWRKRRGRTTLNGGKPVKSLPLSNAEHLKILHDKRVIDYITTVMSN
Summary
Uniprot
H9J2N4
A0A2A4K8P6
A0A2H1V8B5
A0A194QIK3
A0A194QU74
A0A212EML1
+ More
A0A0L7LII1 D6WM55 U4TZ66 A0A1B6CCH8 A0A1Y1MXZ0 A0A1B6GB03 N6TP98 A0A2J7RL47 F4WZ35 A0A195F4N5 A0A026VY17 A0A1B0D3I7 U5ERQ5 A0A195DA32 A0A1B6LYV8 E9IYU9 A0A195CLZ7 A0A067RM07 A0A1L8DYT4 E2A0K2 A0A1L8DYW7 A0A151XEJ7 E2C5D5 A0A2M3Z9A2 A0A232F901 A0A182QYF1 A0A2M4BNN7 A0A2M4ARU8 A0A2M4BNL0 A0A182F2I0 A0A154PM11 A0A2M4BNK1 A0A2M4ADH1 A0A2M4AEF6 K7IP46 W5JDG9 A0A0L7RB39 A0A2M4AAV6 A0A2M4AB17 A0A182LTP9 A0A182NBL1 A0A0N0BHG8 A0A2M4AG49 A0A182KFX5 A0A1W7R8B3 A0A2A3EPM3 A0A182IMM8 A0A182I7K2 A0A182WXN7 A0A182V9A7 A0A1S4GZS6 A0A1S4FYE2 A0A084WC75 J9JZL4 Q16JY8 A0A182Y0I9 A0A182LMH9 A0A182TIV3 A0A2M4CNU8 A0A310SF86 A0A182WGD6 A0A182RHC2 Q5TNA7 A0A2M4AE69 A0A139WGZ4 R4G3P2 A0A182PNH2 A0A3B0K704 B4LSY5 A0A182TBF2 A0A182G5U9 A0A1W4V1E6 A0A1Q3FK64 B3NM13 B4KEP9 B4GSV6 Q29KS6 E0VY54 A0A224XDF2 Q9Y168 C3KGK6 Q9I7L9 B4IFC3 A0A1I8MXY5 A0A1I8P7E8 B3MMU2 A0A0J9TS23 A0A0L0CAC6 A0A1J1IN84 A0A158NE34 B4PBK9 T1PLB5 A0A0P6EQI2 B4JCD2
A0A0L7LII1 D6WM55 U4TZ66 A0A1B6CCH8 A0A1Y1MXZ0 A0A1B6GB03 N6TP98 A0A2J7RL47 F4WZ35 A0A195F4N5 A0A026VY17 A0A1B0D3I7 U5ERQ5 A0A195DA32 A0A1B6LYV8 E9IYU9 A0A195CLZ7 A0A067RM07 A0A1L8DYT4 E2A0K2 A0A1L8DYW7 A0A151XEJ7 E2C5D5 A0A2M3Z9A2 A0A232F901 A0A182QYF1 A0A2M4BNN7 A0A2M4ARU8 A0A2M4BNL0 A0A182F2I0 A0A154PM11 A0A2M4BNK1 A0A2M4ADH1 A0A2M4AEF6 K7IP46 W5JDG9 A0A0L7RB39 A0A2M4AAV6 A0A2M4AB17 A0A182LTP9 A0A182NBL1 A0A0N0BHG8 A0A2M4AG49 A0A182KFX5 A0A1W7R8B3 A0A2A3EPM3 A0A182IMM8 A0A182I7K2 A0A182WXN7 A0A182V9A7 A0A1S4GZS6 A0A1S4FYE2 A0A084WC75 J9JZL4 Q16JY8 A0A182Y0I9 A0A182LMH9 A0A182TIV3 A0A2M4CNU8 A0A310SF86 A0A182WGD6 A0A182RHC2 Q5TNA7 A0A2M4AE69 A0A139WGZ4 R4G3P2 A0A182PNH2 A0A3B0K704 B4LSY5 A0A182TBF2 A0A182G5U9 A0A1W4V1E6 A0A1Q3FK64 B3NM13 B4KEP9 B4GSV6 Q29KS6 E0VY54 A0A224XDF2 Q9Y168 C3KGK6 Q9I7L9 B4IFC3 A0A1I8MXY5 A0A1I8P7E8 B3MMU2 A0A0J9TS23 A0A0L0CAC6 A0A1J1IN84 A0A158NE34 B4PBK9 T1PLB5 A0A0P6EQI2 B4JCD2
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
23537049 28004739 21719571 24508170 30249741 21282665 24845553 20798317 28648823 20075255 20920257 23761445 12364791 24438588 17510324 25244985 20966253 17994087 18057021 26483478 15632085 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 22936249 26108605 21347285 17550304
23537049 28004739 21719571 24508170 30249741 21282665 24845553 20798317 28648823 20075255 20920257 23761445 12364791 24438588 17510324 25244985 20966253 17994087 18057021 26483478 15632085 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 22936249 26108605 21347285 17550304
EMBL
BABH01034447
BABH01034448
NWSH01000050
PCG80183.1
ODYU01001200
SOQ37090.1
+ More
KQ458860 KPJ04760.1 KQ461191 KPJ07096.1 AGBW02013812 OWR42723.1 JTDY01000935 KOB75373.1 KQ971343 EFA04227.1 KB631676 ERL85318.1 GEDC01026141 JAS11157.1 GEZM01017645 GEZM01017642 GEZM01017640 GEZM01017633 JAV90524.1 GECZ01010151 JAS59618.1 APGK01057250 APGK01057251 APGK01057252 KB741279 ENN71045.1 NEVH01002699 PNF41566.1 GL888465 EGI60538.1 KQ981820 KYN35137.1 KK107578 QOIP01000002 EZA48688.1 RLU25851.1 AJVK01010992 AJVK01010993 AJVK01010994 GANO01002711 JAB57160.1 KQ981082 KYN09722.1 GEBQ01011110 JAT28867.1 GL767051 EFZ14262.1 KQ977600 KYN01492.1 KK852595 KDR20632.1 GFDF01002466 JAV11618.1 GL435626 EFN72905.1 GFDF01002465 JAV11619.1 KQ982254 KYQ58789.1 GL452770 EFN76868.1 GGFM01004352 MBW25103.1 NNAY01000623 OXU27331.1 AXCN02000676 GGFJ01005524 MBW54665.1 GGFK01010205 MBW43526.1 GGFJ01005526 MBW54667.1 KQ434977 KZC12915.1 GGFJ01005525 MBW54666.1 GGFK01005515 MBW38836.1 GGFK01005781 MBW39102.1 ADMH02001853 ETN60859.1 KQ414617 KOC68058.1 GGFK01004600 MBW37921.1 GGFK01004656 MBW37977.1 AXCM01003792 KQ435752 KOX76218.1 GGFK01006444 MBW39765.1 GEHC01000246 JAV47399.1 KZ288203 PBC33454.1 APCN01003414 AAAB01008984 ATLV01022579 KE525333 KFB47819.1 ABLF02037432 CH477986 EAT34595.1 GGFL01002410 MBW66588.1 KQ775154 OAD52278.1 EAL39003.2 GGFK01005765 MBW39086.1 KYB27268.1 GAHY01002050 JAA75460.1 OUUW01000006 SPP81799.1 CH940649 EDW64896.1 KRF81930.1 JXUM01146613 JXUM01146614 KQ570209 KXJ68422.1 GFDL01007044 JAV28001.1 CH954179 EDV54549.1 CH933807 EDW12949.1 CH479189 EDW25465.1 CH379061 EAL33099.1 DS235843 EEB18310.1 GFTR01005951 JAW10475.1 AF145599 AE014134 AAD38574.1 AAN11076.2 AGB93161.1 BT082071 ACP28232.1 AAG22446.2 AGB93163.1 CH480833 EDW46345.1 CH902620 EDV30967.1 CM002910 KMY91165.1 JRES01000682 KNC29190.1 CVRI01000055 CRL01624.1 ADTU01012727 ADTU01012728 CM000158 EDW90523.1 KA646581 KA649562 AFP64191.1 GDIQ01059270 JAN35467.1 CH916368 EDW04165.1
KQ458860 KPJ04760.1 KQ461191 KPJ07096.1 AGBW02013812 OWR42723.1 JTDY01000935 KOB75373.1 KQ971343 EFA04227.1 KB631676 ERL85318.1 GEDC01026141 JAS11157.1 GEZM01017645 GEZM01017642 GEZM01017640 GEZM01017633 JAV90524.1 GECZ01010151 JAS59618.1 APGK01057250 APGK01057251 APGK01057252 KB741279 ENN71045.1 NEVH01002699 PNF41566.1 GL888465 EGI60538.1 KQ981820 KYN35137.1 KK107578 QOIP01000002 EZA48688.1 RLU25851.1 AJVK01010992 AJVK01010993 AJVK01010994 GANO01002711 JAB57160.1 KQ981082 KYN09722.1 GEBQ01011110 JAT28867.1 GL767051 EFZ14262.1 KQ977600 KYN01492.1 KK852595 KDR20632.1 GFDF01002466 JAV11618.1 GL435626 EFN72905.1 GFDF01002465 JAV11619.1 KQ982254 KYQ58789.1 GL452770 EFN76868.1 GGFM01004352 MBW25103.1 NNAY01000623 OXU27331.1 AXCN02000676 GGFJ01005524 MBW54665.1 GGFK01010205 MBW43526.1 GGFJ01005526 MBW54667.1 KQ434977 KZC12915.1 GGFJ01005525 MBW54666.1 GGFK01005515 MBW38836.1 GGFK01005781 MBW39102.1 ADMH02001853 ETN60859.1 KQ414617 KOC68058.1 GGFK01004600 MBW37921.1 GGFK01004656 MBW37977.1 AXCM01003792 KQ435752 KOX76218.1 GGFK01006444 MBW39765.1 GEHC01000246 JAV47399.1 KZ288203 PBC33454.1 APCN01003414 AAAB01008984 ATLV01022579 KE525333 KFB47819.1 ABLF02037432 CH477986 EAT34595.1 GGFL01002410 MBW66588.1 KQ775154 OAD52278.1 EAL39003.2 GGFK01005765 MBW39086.1 KYB27268.1 GAHY01002050 JAA75460.1 OUUW01000006 SPP81799.1 CH940649 EDW64896.1 KRF81930.1 JXUM01146613 JXUM01146614 KQ570209 KXJ68422.1 GFDL01007044 JAV28001.1 CH954179 EDV54549.1 CH933807 EDW12949.1 CH479189 EDW25465.1 CH379061 EAL33099.1 DS235843 EEB18310.1 GFTR01005951 JAW10475.1 AF145599 AE014134 AAD38574.1 AAN11076.2 AGB93161.1 BT082071 ACP28232.1 AAG22446.2 AGB93163.1 CH480833 EDW46345.1 CH902620 EDV30967.1 CM002910 KMY91165.1 JRES01000682 KNC29190.1 CVRI01000055 CRL01624.1 ADTU01012727 ADTU01012728 CM000158 EDW90523.1 KA646581 KA649562 AFP64191.1 GDIQ01059270 JAN35467.1 CH916368 EDW04165.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000007266 UP000030742 UP000019118 UP000235965 UP000007755 UP000078541 UP000053097 UP000279307 UP000092462 UP000078492 UP000078542 UP000027135 UP000000311 UP000075809 UP000008237 UP000215335 UP000075886 UP000069272 UP000076502 UP000002358 UP000000673 UP000053825 UP000075883 UP000075884 UP000053105 UP000075881 UP000242457 UP000075880 UP000075840 UP000076407 UP000075903 UP000030765 UP000007819 UP000008820 UP000076408 UP000075882 UP000075902 UP000075920 UP000075900 UP000007062 UP000075885 UP000268350 UP000008792 UP000075901 UP000069940 UP000249989 UP000192221 UP000008711 UP000009192 UP000008744 UP000001819 UP000009046 UP000000803 UP000001292 UP000095301 UP000095300 UP000007801 UP000037069 UP000183832 UP000005205 UP000002282 UP000001070
UP000007266 UP000030742 UP000019118 UP000235965 UP000007755 UP000078541 UP000053097 UP000279307 UP000092462 UP000078492 UP000078542 UP000027135 UP000000311 UP000075809 UP000008237 UP000215335 UP000075886 UP000069272 UP000076502 UP000002358 UP000000673 UP000053825 UP000075883 UP000075884 UP000053105 UP000075881 UP000242457 UP000075880 UP000075840 UP000076407 UP000075903 UP000030765 UP000007819 UP000008820 UP000076408 UP000075882 UP000075902 UP000075920 UP000075900 UP000007062 UP000075885 UP000268350 UP000008792 UP000075901 UP000069940 UP000249989 UP000192221 UP000008711 UP000009192 UP000008744 UP000001819 UP000009046 UP000000803 UP000001292 UP000095301 UP000095300 UP000007801 UP000037069 UP000183832 UP000005205 UP000002282 UP000001070
Interpro
Gene 3D
ProteinModelPortal
H9J2N4
A0A2A4K8P6
A0A2H1V8B5
A0A194QIK3
A0A194QU74
A0A212EML1
+ More
A0A0L7LII1 D6WM55 U4TZ66 A0A1B6CCH8 A0A1Y1MXZ0 A0A1B6GB03 N6TP98 A0A2J7RL47 F4WZ35 A0A195F4N5 A0A026VY17 A0A1B0D3I7 U5ERQ5 A0A195DA32 A0A1B6LYV8 E9IYU9 A0A195CLZ7 A0A067RM07 A0A1L8DYT4 E2A0K2 A0A1L8DYW7 A0A151XEJ7 E2C5D5 A0A2M3Z9A2 A0A232F901 A0A182QYF1 A0A2M4BNN7 A0A2M4ARU8 A0A2M4BNL0 A0A182F2I0 A0A154PM11 A0A2M4BNK1 A0A2M4ADH1 A0A2M4AEF6 K7IP46 W5JDG9 A0A0L7RB39 A0A2M4AAV6 A0A2M4AB17 A0A182LTP9 A0A182NBL1 A0A0N0BHG8 A0A2M4AG49 A0A182KFX5 A0A1W7R8B3 A0A2A3EPM3 A0A182IMM8 A0A182I7K2 A0A182WXN7 A0A182V9A7 A0A1S4GZS6 A0A1S4FYE2 A0A084WC75 J9JZL4 Q16JY8 A0A182Y0I9 A0A182LMH9 A0A182TIV3 A0A2M4CNU8 A0A310SF86 A0A182WGD6 A0A182RHC2 Q5TNA7 A0A2M4AE69 A0A139WGZ4 R4G3P2 A0A182PNH2 A0A3B0K704 B4LSY5 A0A182TBF2 A0A182G5U9 A0A1W4V1E6 A0A1Q3FK64 B3NM13 B4KEP9 B4GSV6 Q29KS6 E0VY54 A0A224XDF2 Q9Y168 C3KGK6 Q9I7L9 B4IFC3 A0A1I8MXY5 A0A1I8P7E8 B3MMU2 A0A0J9TS23 A0A0L0CAC6 A0A1J1IN84 A0A158NE34 B4PBK9 T1PLB5 A0A0P6EQI2 B4JCD2
A0A0L7LII1 D6WM55 U4TZ66 A0A1B6CCH8 A0A1Y1MXZ0 A0A1B6GB03 N6TP98 A0A2J7RL47 F4WZ35 A0A195F4N5 A0A026VY17 A0A1B0D3I7 U5ERQ5 A0A195DA32 A0A1B6LYV8 E9IYU9 A0A195CLZ7 A0A067RM07 A0A1L8DYT4 E2A0K2 A0A1L8DYW7 A0A151XEJ7 E2C5D5 A0A2M3Z9A2 A0A232F901 A0A182QYF1 A0A2M4BNN7 A0A2M4ARU8 A0A2M4BNL0 A0A182F2I0 A0A154PM11 A0A2M4BNK1 A0A2M4ADH1 A0A2M4AEF6 K7IP46 W5JDG9 A0A0L7RB39 A0A2M4AAV6 A0A2M4AB17 A0A182LTP9 A0A182NBL1 A0A0N0BHG8 A0A2M4AG49 A0A182KFX5 A0A1W7R8B3 A0A2A3EPM3 A0A182IMM8 A0A182I7K2 A0A182WXN7 A0A182V9A7 A0A1S4GZS6 A0A1S4FYE2 A0A084WC75 J9JZL4 Q16JY8 A0A182Y0I9 A0A182LMH9 A0A182TIV3 A0A2M4CNU8 A0A310SF86 A0A182WGD6 A0A182RHC2 Q5TNA7 A0A2M4AE69 A0A139WGZ4 R4G3P2 A0A182PNH2 A0A3B0K704 B4LSY5 A0A182TBF2 A0A182G5U9 A0A1W4V1E6 A0A1Q3FK64 B3NM13 B4KEP9 B4GSV6 Q29KS6 E0VY54 A0A224XDF2 Q9Y168 C3KGK6 Q9I7L9 B4IFC3 A0A1I8MXY5 A0A1I8P7E8 B3MMU2 A0A0J9TS23 A0A0L0CAC6 A0A1J1IN84 A0A158NE34 B4PBK9 T1PLB5 A0A0P6EQI2 B4JCD2
PDB
6MTW
E-value=4.19909e-96,
Score=897
Ontologies
PATHWAY
GO
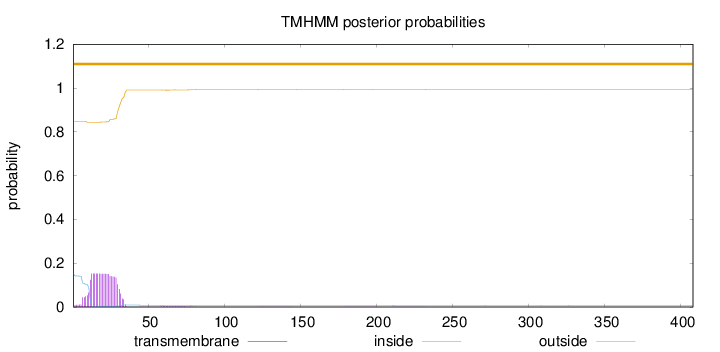
Topology
SignalP
Position: 1 - 23,
Likelihood: 0.862265
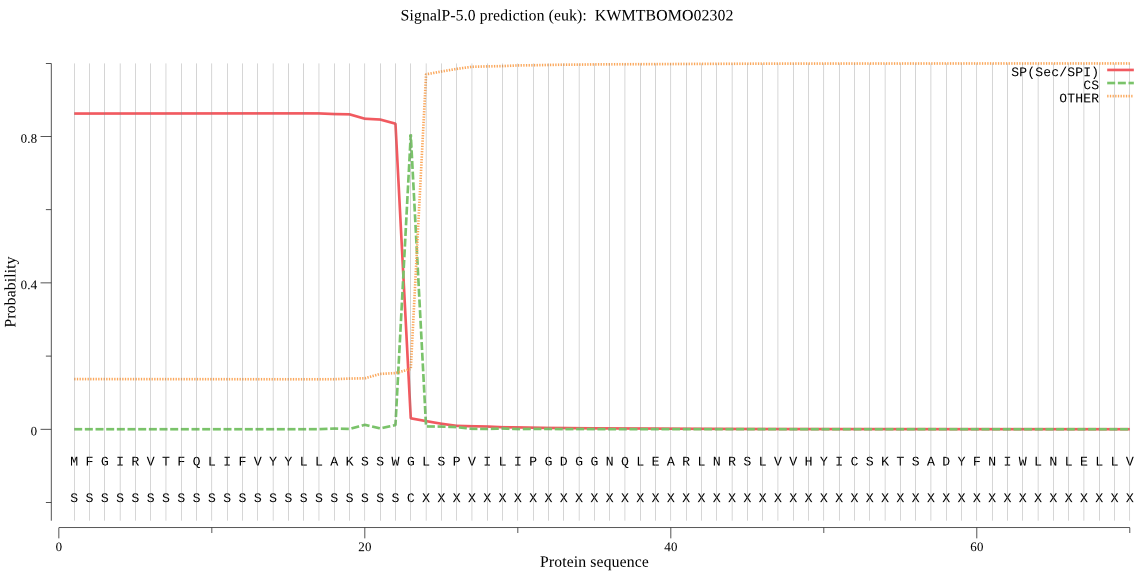
Length:
408
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.37153000000001
Exp number, first 60 AAs:
3.29338
Total prob of N-in:
0.15145
outside
1 - 408
Population Genetic Test Statistics
Pi
213.580832
Theta
2.249615
Tajima's D
0.158913
CLR
11.398088
CSRT
0.456277186140693
Interpretation
Uncertain
