Gene
KWMTBOMO02301
Pre Gene Modal
BGIBMGA003782
Annotation
PREDICTED:_TBC1_domain_family_member_20?_partial_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.648 Nuclear Reliability : 1.54
Sequence
CDS
ATGGAATCCAATAATAAAGACAGTGAAGTTATAGATAAAAGTGTTATGAATGGTGAATGTTCACCAGCAGAGAGTATTCATCCGACAAATTTGAATAAAACGAAAAATATTGATGTCATTTCGACACCCTCCGAATTAAAATTCGATAATGAGCCGGAGGAGGAAGACCCCGAGATTGTAGAAAAGAGGAAAGAGATTGAGGAATGTCTCTCTGATGTCGATTCTGTTGATGTGGAAAAATGGAAGAATCTAGCCAGAAGCAAAGGAGGACTTATCTGTGATGAATACAGAAGAAGAATTTGGCCTCTGCTTGTTGGTGTTACAAAAGAGGAAATGACTGAAGCTCCATCATTGGATGAGTTATCAACGCATCCAGAATATAACCAGGTGGTGCTAGATGTGAACAGGTCTTTGAAAAGATTTCCTCCTGGCATTCCATACGAACAAAGAGTGGCCCTTCAGGACCAGCTGACAGTATTAATACTCAGGGTCATCATAAAATATCCACATTTAAAATATTACCAGGGTTACCATGACGTGGCGATCACCCTGCTCTTGGTGTGCGGAGACCGTGCCTCGTTCCCGCTGCTGTGCCGCCTGTCCTACGGCGCCGGGGCTCCGCTGGCGCCCTTCATGCAGCTCACGATGCAGCCCACGCAGCATCTGCTCAATTACATGCTGCCAGTCGTGGGCAGAGCCAATACCAAACTGGCCAATAGACTTGAAGAGGCGGGTGTGGGCACGATGTTCGCGCTGCCGTGGTACCTGACGTGGTTCGGGCACAGCCTGGGCCGGTACGCGCTCGTCGTGCGCCTCTACGACTACTTCCTGTGCGCGCCGCCGCTCTTCCCCGTGTACGTCACCGCCGCCGTCGTGCTGTGCAGAGCCGATGAAGTGTTTGAATGTGAACTGGACATGGCTATGCTGCATTGTTTACTGTCAAAGTTACCGGATGACTTGCCCTTCGAAGACATCTTGGTGACTGCCCAGGAGCTCTACGAGAACAACGACCCCCTCGACCTCGAGGACGAAGTCGTCGCACTCGAGCAGAAAGAGTGA
Protein
MESNNKDSEVIDKSVMNGECSPAESIHPTNLNKTKNIDVISTPSELKFDNEPEEEDPEIVEKRKEIEECLSDVDSVDVEKWKNLARSKGGLICDEYRRRIWPLLVGVTKEEMTEAPSLDELSTHPEYNQVVLDVNRSLKRFPPGIPYEQRVALQDQLTVLILRVIIKYPHLKYYQGYHDVAITLLLVCGDRASFPLLCRLSYGAGAPLAPFMQLTMQPTQHLLNYMLPVVGRANTKLANRLEEAGVGTMFALPWYLTWFGHSLGRYALVVRLYDYFLCAPPLFPVYVTAAVVLCRADEVFECELDMAMLHCLLSKLPDDLPFEDILVTAQELYENNDPLDLEDEVVALEQKE
Summary
Uniprot
H9J2P5
A0A212EMK6
A0A2A4K940
A0A194QNK7
A0A194QH65
A0A0L7LIW4
+ More
U5EPB7 Q7Q4A6 A0A182JYL8 A0A182RMC2 A0A182Y3D8 A0A182MB07 Q17L28 A0A1L8DLR7 A0A182PAV7 A0A182XL88 A0A182QKK8 A0A182ICD9 A0A182VHD8 A0A182L0S7 A0A182U4Z2 A0A1S4GYN1 A0A182W6M4 A0A2M4BS83 A0A182NTT0 A0A2M4AQ61 A0A084WBV2 W5J3J4 A0A182H5U7 A0A182F4Z6 A0A182H8C0 A0A0T6AUY3 A0A1Q3FX72 A0A1Q3FXF2 A0A336LQX9 A0A1B0CZR8 A0A1Y1NEC9 A0A0K8TR81 U4TUT9 A0A1B6CZ04 A0A2J7QCU1 A0A1I8NG41 A0A0L0BQ76 A0A182SD61 N6TRX3 A0A1J1I456 B4GX92 A0A0K8U2F8 B4JPR0 A0A1B6GG14 A0A067RA51 A0A1I8P6J4 A0A0A1X9H8 A0A1A9WXE1 A0A1S4JPU6 B5DTF8 B0WQ80 A0A1B6L9V0 A0A182JIW8 B4LQZ0 D6WM56 A0A034WR29 A0A146L848 W8ASY3 A0A1A9Z446 A0A1A9UYR9 A0A0M4E1E5 A0A0V0GCC1 A0A224XH78 A0A3B0KBQ0 B4KJ12 D3TQC2 T1HTI0 B4NNX8 A0A2R7WWA3 R4G8Y8 A0A069DU99 A0A0P8Y7W6 B3N4P2 B4NZG0 A0A1W4UQQ4 A0A0R1DN96 A0A1A9YJW4 A0A1B0BQG1 Q9W5H8 B4NV68 B4ILL3 A0A0B4KEC2 A0A0J9TTM2 E0VY55 A0A0B4KEH8 A0A1S3ICT5 A0A1S3IDY9 A0A067ZT82 A0A0K2TK69 A0A2I4B8M8
U5EPB7 Q7Q4A6 A0A182JYL8 A0A182RMC2 A0A182Y3D8 A0A182MB07 Q17L28 A0A1L8DLR7 A0A182PAV7 A0A182XL88 A0A182QKK8 A0A182ICD9 A0A182VHD8 A0A182L0S7 A0A182U4Z2 A0A1S4GYN1 A0A182W6M4 A0A2M4BS83 A0A182NTT0 A0A2M4AQ61 A0A084WBV2 W5J3J4 A0A182H5U7 A0A182F4Z6 A0A182H8C0 A0A0T6AUY3 A0A1Q3FX72 A0A1Q3FXF2 A0A336LQX9 A0A1B0CZR8 A0A1Y1NEC9 A0A0K8TR81 U4TUT9 A0A1B6CZ04 A0A2J7QCU1 A0A1I8NG41 A0A0L0BQ76 A0A182SD61 N6TRX3 A0A1J1I456 B4GX92 A0A0K8U2F8 B4JPR0 A0A1B6GG14 A0A067RA51 A0A1I8P6J4 A0A0A1X9H8 A0A1A9WXE1 A0A1S4JPU6 B5DTF8 B0WQ80 A0A1B6L9V0 A0A182JIW8 B4LQZ0 D6WM56 A0A034WR29 A0A146L848 W8ASY3 A0A1A9Z446 A0A1A9UYR9 A0A0M4E1E5 A0A0V0GCC1 A0A224XH78 A0A3B0KBQ0 B4KJ12 D3TQC2 T1HTI0 B4NNX8 A0A2R7WWA3 R4G8Y8 A0A069DU99 A0A0P8Y7W6 B3N4P2 B4NZG0 A0A1W4UQQ4 A0A0R1DN96 A0A1A9YJW4 A0A1B0BQG1 Q9W5H8 B4NV68 B4ILL3 A0A0B4KEC2 A0A0J9TTM2 E0VY55 A0A0B4KEH8 A0A1S3ICT5 A0A1S3IDY9 A0A067ZT82 A0A0K2TK69 A0A2I4B8M8
Pubmed
19121390
22118469
26354079
26227816
12364791
25244985
+ More
17510324 20966253 24438588 20920257 23761445 26483478 28004739 26369729 23537049 25315136 26108605 17994087 24845553 25830018 15632085 18362917 19820115 25348373 26823975 24495485 20353571 26334808 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20566863
17510324 20966253 24438588 20920257 23761445 26483478 28004739 26369729 23537049 25315136 26108605 17994087 24845553 25830018 15632085 18362917 19820115 25348373 26823975 24495485 20353571 26334808 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20566863
EMBL
BABH01034447
AGBW02013812
OWR42722.1
NWSH01000050
PCG80182.1
KQ461191
+ More
KPJ07097.1 KQ458860 KPJ04759.1 JTDY01000935 KOB75370.1 GANO01003748 JAB56123.1 AAAB01008964 EAA12204.4 AXCM01002003 CH477219 EAT47398.1 GFDF01006671 JAV07413.1 AXCN02000803 APCN01000797 GGFJ01006702 MBW55843.1 GGFK01009589 MBW42910.1 ATLV01022472 KE525332 KFB47696.1 ADMH02002110 ETN58917.1 JXUM01026673 JXUM01026674 KQ560764 KXJ80972.1 JXUM01118080 KQ566147 KXJ70365.1 LJIG01022744 KRT78943.1 GFDL01002835 JAV32210.1 GFDL01002869 JAV32176.1 UFQT01000055 SSX19093.1 AJVK01009695 AJVK01009696 AJVK01009697 GEZM01009504 JAV94546.1 GDAI01000721 JAI16882.1 KB631676 ERL85319.1 GEDC01026199 GEDC01024939 GEDC01018723 JAS11099.1 JAS12359.1 JAS18575.1 NEVH01015876 PNF26397.1 JRES01001533 KNC22212.1 APGK01057255 APGK01057256 APGK01057257 KB741279 ENN71046.1 CVRI01000038 CRK94380.1 CH479195 EDW27203.1 GDHF01031380 JAI20934.1 CH916372 EDV98890.1 GECZ01008506 JAS61263.1 KK852595 KDR20624.1 GBXI01010613 GBXI01006917 JAD03679.1 JAD07375.1 CH475874 EDY71168.1 DS232036 EDS32740.1 GEBQ01019504 JAT20473.1 CH940649 EDW64529.2 KQ971343 EFA03356.2 GAKP01001853 JAC57099.1 GDHC01014086 JAQ04543.1 GAMC01017258 JAB89297.1 CP012523 ALC39166.1 GECL01000412 JAP05712.1 GFTR01006042 JAW10384.1 OUUW01000010 SPP85590.1 CH933807 EDW13525.1 EZ423624 ADD19900.1 ACPB03016020 CH964289 EDW86067.1 KK855763 PTY23822.1 GAHY01000147 JAA77363.1 GBGD01001607 JAC87282.1 CH902628 KPU75343.1 CH954177 EDV59996.1 CM000157 EDW89873.1 KRJ98762.1 JXJN01018612 AY113520 AE013599 AAM29525.1 EAA45988.2 CH986366 CM002911 EDX15943.1 KMY91564.1 CH480875 EDW54667.1 AGB93236.1 KMY91565.1 DS235843 EEB18311.1 AGB93237.1 KF039720 AHX25870.1 HACA01008530 CDW25891.1
KPJ07097.1 KQ458860 KPJ04759.1 JTDY01000935 KOB75370.1 GANO01003748 JAB56123.1 AAAB01008964 EAA12204.4 AXCM01002003 CH477219 EAT47398.1 GFDF01006671 JAV07413.1 AXCN02000803 APCN01000797 GGFJ01006702 MBW55843.1 GGFK01009589 MBW42910.1 ATLV01022472 KE525332 KFB47696.1 ADMH02002110 ETN58917.1 JXUM01026673 JXUM01026674 KQ560764 KXJ80972.1 JXUM01118080 KQ566147 KXJ70365.1 LJIG01022744 KRT78943.1 GFDL01002835 JAV32210.1 GFDL01002869 JAV32176.1 UFQT01000055 SSX19093.1 AJVK01009695 AJVK01009696 AJVK01009697 GEZM01009504 JAV94546.1 GDAI01000721 JAI16882.1 KB631676 ERL85319.1 GEDC01026199 GEDC01024939 GEDC01018723 JAS11099.1 JAS12359.1 JAS18575.1 NEVH01015876 PNF26397.1 JRES01001533 KNC22212.1 APGK01057255 APGK01057256 APGK01057257 KB741279 ENN71046.1 CVRI01000038 CRK94380.1 CH479195 EDW27203.1 GDHF01031380 JAI20934.1 CH916372 EDV98890.1 GECZ01008506 JAS61263.1 KK852595 KDR20624.1 GBXI01010613 GBXI01006917 JAD03679.1 JAD07375.1 CH475874 EDY71168.1 DS232036 EDS32740.1 GEBQ01019504 JAT20473.1 CH940649 EDW64529.2 KQ971343 EFA03356.2 GAKP01001853 JAC57099.1 GDHC01014086 JAQ04543.1 GAMC01017258 JAB89297.1 CP012523 ALC39166.1 GECL01000412 JAP05712.1 GFTR01006042 JAW10384.1 OUUW01000010 SPP85590.1 CH933807 EDW13525.1 EZ423624 ADD19900.1 ACPB03016020 CH964289 EDW86067.1 KK855763 PTY23822.1 GAHY01000147 JAA77363.1 GBGD01001607 JAC87282.1 CH902628 KPU75343.1 CH954177 EDV59996.1 CM000157 EDW89873.1 KRJ98762.1 JXJN01018612 AY113520 AE013599 AAM29525.1 EAA45988.2 CH986366 CM002911 EDX15943.1 KMY91564.1 CH480875 EDW54667.1 AGB93236.1 KMY91565.1 DS235843 EEB18311.1 AGB93237.1 KF039720 AHX25870.1 HACA01008530 CDW25891.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000053268
UP000037510
+ More
UP000007062 UP000075881 UP000075900 UP000076408 UP000075883 UP000008820 UP000075885 UP000076407 UP000075886 UP000075840 UP000075903 UP000075882 UP000075902 UP000075920 UP000075884 UP000030765 UP000000673 UP000069940 UP000249989 UP000069272 UP000092462 UP000030742 UP000235965 UP000095301 UP000037069 UP000075901 UP000019118 UP000183832 UP000008744 UP000001070 UP000027135 UP000095300 UP000091820 UP000001819 UP000002320 UP000075880 UP000008792 UP000007266 UP000092445 UP000078200 UP000092553 UP000268350 UP000009192 UP000015103 UP000007798 UP000007801 UP000008711 UP000002282 UP000192221 UP000092443 UP000092460 UP000000803 UP000000304 UP000001292 UP000009046 UP000085678 UP000192220
UP000007062 UP000075881 UP000075900 UP000076408 UP000075883 UP000008820 UP000075885 UP000076407 UP000075886 UP000075840 UP000075903 UP000075882 UP000075902 UP000075920 UP000075884 UP000030765 UP000000673 UP000069940 UP000249989 UP000069272 UP000092462 UP000030742 UP000235965 UP000095301 UP000037069 UP000075901 UP000019118 UP000183832 UP000008744 UP000001070 UP000027135 UP000095300 UP000091820 UP000001819 UP000002320 UP000075880 UP000008792 UP000007266 UP000092445 UP000078200 UP000092553 UP000268350 UP000009192 UP000015103 UP000007798 UP000007801 UP000008711 UP000002282 UP000192221 UP000092443 UP000092460 UP000000803 UP000000304 UP000001292 UP000009046 UP000085678 UP000192220
Pfam
PF00566 RabGAP-TBC
SUPFAM
SSF47923
SSF47923
ProteinModelPortal
H9J2P5
A0A212EMK6
A0A2A4K940
A0A194QNK7
A0A194QH65
A0A0L7LIW4
+ More
U5EPB7 Q7Q4A6 A0A182JYL8 A0A182RMC2 A0A182Y3D8 A0A182MB07 Q17L28 A0A1L8DLR7 A0A182PAV7 A0A182XL88 A0A182QKK8 A0A182ICD9 A0A182VHD8 A0A182L0S7 A0A182U4Z2 A0A1S4GYN1 A0A182W6M4 A0A2M4BS83 A0A182NTT0 A0A2M4AQ61 A0A084WBV2 W5J3J4 A0A182H5U7 A0A182F4Z6 A0A182H8C0 A0A0T6AUY3 A0A1Q3FX72 A0A1Q3FXF2 A0A336LQX9 A0A1B0CZR8 A0A1Y1NEC9 A0A0K8TR81 U4TUT9 A0A1B6CZ04 A0A2J7QCU1 A0A1I8NG41 A0A0L0BQ76 A0A182SD61 N6TRX3 A0A1J1I456 B4GX92 A0A0K8U2F8 B4JPR0 A0A1B6GG14 A0A067RA51 A0A1I8P6J4 A0A0A1X9H8 A0A1A9WXE1 A0A1S4JPU6 B5DTF8 B0WQ80 A0A1B6L9V0 A0A182JIW8 B4LQZ0 D6WM56 A0A034WR29 A0A146L848 W8ASY3 A0A1A9Z446 A0A1A9UYR9 A0A0M4E1E5 A0A0V0GCC1 A0A224XH78 A0A3B0KBQ0 B4KJ12 D3TQC2 T1HTI0 B4NNX8 A0A2R7WWA3 R4G8Y8 A0A069DU99 A0A0P8Y7W6 B3N4P2 B4NZG0 A0A1W4UQQ4 A0A0R1DN96 A0A1A9YJW4 A0A1B0BQG1 Q9W5H8 B4NV68 B4ILL3 A0A0B4KEC2 A0A0J9TTM2 E0VY55 A0A0B4KEH8 A0A1S3ICT5 A0A1S3IDY9 A0A067ZT82 A0A0K2TK69 A0A2I4B8M8
U5EPB7 Q7Q4A6 A0A182JYL8 A0A182RMC2 A0A182Y3D8 A0A182MB07 Q17L28 A0A1L8DLR7 A0A182PAV7 A0A182XL88 A0A182QKK8 A0A182ICD9 A0A182VHD8 A0A182L0S7 A0A182U4Z2 A0A1S4GYN1 A0A182W6M4 A0A2M4BS83 A0A182NTT0 A0A2M4AQ61 A0A084WBV2 W5J3J4 A0A182H5U7 A0A182F4Z6 A0A182H8C0 A0A0T6AUY3 A0A1Q3FX72 A0A1Q3FXF2 A0A336LQX9 A0A1B0CZR8 A0A1Y1NEC9 A0A0K8TR81 U4TUT9 A0A1B6CZ04 A0A2J7QCU1 A0A1I8NG41 A0A0L0BQ76 A0A182SD61 N6TRX3 A0A1J1I456 B4GX92 A0A0K8U2F8 B4JPR0 A0A1B6GG14 A0A067RA51 A0A1I8P6J4 A0A0A1X9H8 A0A1A9WXE1 A0A1S4JPU6 B5DTF8 B0WQ80 A0A1B6L9V0 A0A182JIW8 B4LQZ0 D6WM56 A0A034WR29 A0A146L848 W8ASY3 A0A1A9Z446 A0A1A9UYR9 A0A0M4E1E5 A0A0V0GCC1 A0A224XH78 A0A3B0KBQ0 B4KJ12 D3TQC2 T1HTI0 B4NNX8 A0A2R7WWA3 R4G8Y8 A0A069DU99 A0A0P8Y7W6 B3N4P2 B4NZG0 A0A1W4UQQ4 A0A0R1DN96 A0A1A9YJW4 A0A1B0BQG1 Q9W5H8 B4NV68 B4ILL3 A0A0B4KEC2 A0A0J9TTM2 E0VY55 A0A0B4KEH8 A0A1S3ICT5 A0A1S3IDY9 A0A067ZT82 A0A0K2TK69 A0A2I4B8M8
PDB
4HLQ
E-value=3.60939e-61,
Score=595
Ontologies
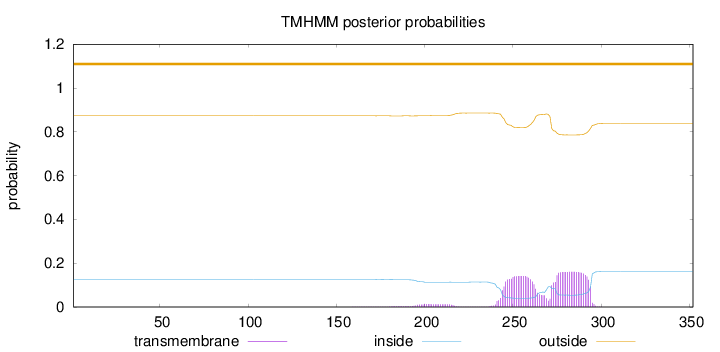
Topology
Length:
352
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.71598
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12673
outside
1 - 352
Population Genetic Test Statistics
Pi
259.524287
Theta
149.527127
Tajima's D
1.987021
CLR
0.658224
CSRT
0.879906004699765
Interpretation
Uncertain
