Gene
KWMTBOMO02297
Pre Gene Modal
BGIBMGA003778
Annotation
PREDICTED:_uncharacterized_protein_C630.12-like_[Bombyx_mori]
Full name
DNA polymerase
Location in the cell
PlasmaMembrane Reliability : 3.484
Sequence
CDS
ATGTATCTTAAAAGAAGTTCAACAGTGAAGTACTTCGCAGCCGCAATAATAGGATGTTTACTATATAGCGAATGGTTAATATATTCGATGCAGGCTTGGTATTGGAGAACCCTGGAGTGCGTCGAATACGACTCATCTTGCATCAAGATACTGTTCGTAGCAGATCCTCAGATACAGGGCGACATAGCGGTGCCGGCACCTTTTAGCTATCCAATTAATTGGGATAGTGATCGTTACCTTAAGTCAACATTTAGAGTTGTTATACAGCACTTCAGGCCAGATGTTATTGTGTACATGGGTGACCTCATGGATGAAGGTTCCATTTCCACCTTGAAACAATACCATAGCTATGTAAAAAGACTGTCTGATATATTTGAAGTTGATTATCCTATAGTGCAAATTTGGATACCAGGTGATAATGATATAGGTGGTGAGAATGAACCGATTAAGAAAGATAAAATTGAAGAGTTCAACGAAGTCTACCAACAACCAGAAACTGTGACATTCCGGAATATAACATTTTACAAAGCCAATGGTATAACGAACAGTTTCCCAAAGAAACTTAAGGGGAATGGCAATGAATACAGGATTGTGGTCTCACATTATCCTATTACATACAGACATGCTTTTGGTCATGAAGTGAATAATGAAATAAATCCGGATATATACTTCTGCGCTCACGAGCACGAGGCAAAATATGTCAGACAGAGTCGAAAACTGACCAATCGAGACTCACACATGCTAACCTCAGAGAGCGCCGTCCTGAACGTGTCAACCAAAGACGATCATTTGTATGAAATTTACGTTCCTACGTGTTCATATAGGATGGGCACAGATCAAATTGGTTATGGTGCTGCCATACTTGAGAACAACAATCAGAACTTGCGTTACACAGTGTTCTGGTCGTCACGAAGGTTTCCCTACTTATTCATTTATTTAGTAACCTTCATCATACTGCAGATCTACTTTGTAATATACTGCGCGGCTTGGGTCAGTTACAGGAAGCCAACGATGAACTGGGCCACTGATAAAGTGCCATTATTGGACAGAGTATAA
Protein
MYLKRSSTVKYFAAAIIGCLLYSEWLIYSMQAWYWRTLECVEYDSSCIKILFVADPQIQGDIAVPAPFSYPINWDSDRYLKSTFRVVIQHFRPDVIVYMGDLMDEGSISTLKQYHSYVKRLSDIFEVDYPIVQIWIPGDNDIGGENEPIKKDKIEEFNEVYQQPETVTFRNITFYKANGITNSFPKKLKGNGNEYRIVVSHYPITYRHAFGHEVNNEINPDIYFCAHEHEAKYVRQSRKLTNRDSHMLTSESAVLNVSTKDDHLYEIYVPTCSYRMGTDQIGYGAAILENNNQNLRYTVFWSSRRFPYLFIYLVTFIILQIYFVIYCAAWVSYRKPTMNWATDKVPLLDRV
Summary
Description
DNA polymerase that functions in several pathways of DNA repair. Involved in base excision repair (BER) responsible for repair of lesions that give rise to abasic (AP) sites in DNA. Also contributes to DNA double-strand break repair by non-homologous end joining and homologous recombination. Has both template-dependent and template-independent (terminal transferase) DNA polymerase activities. Has also a 5'-deoxyribose-5-phosphate lyase (dRP lyase) activity.
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Similarity
Belongs to the DNA polymerase type-X family.
Uniprot
H9J2P1
H9JXP7
A0A2A4K919
A0A212EMK8
A0A194QMP4
A0A194QNK3
+ More
A0A0L7LTI5 A0A0T6B8U5 A0A2A3E6L6 A0A087ZR85 A0A1L8DWE6 A0A1L8DWU5 E2BFY9 A0A2J7Q2E3 A0A067R9P4 A0A0L7QQK1 A0A195CUT9 A0A2J7Q2F7 F4WC33 E9IKB1 A0A0M9AAK4 A0A026WWD5 E2AH74 A0A154P5E5 A0A182PCI1 A0A232EPN5 A0A1B6CQ96 N6ULY0 A0A182M1I5 D6WH01 A0A195DLQ4 A0A195BKF4 A0A195FR16 A0A182WA52 A0A182QND2 A0A182YIM1 A0A182R7Q2 A0A1W4W9F7 A0A131XBC3 A0A158NAQ8 A0A310SJQ9 B0W1N3 Q7Q0U8 A0A182HQV4 A0A182UEW1 A0A182XLF4 A0A1S4J383 A0A2R5L7J8 A0A1Y1K5A4 A0A182NPW2 A0A182VB61 A0A0J7KSX3 A0A1Q3FJX7 A0A1Q3FJW6 L7LZP2 A0A2H8TRQ1 A0A182KJL0 A0A2J7Q2E5 A0A1J1J8A2 A0A084WGA6 B7PTK5 A0A131Y4D2 A0A182K4Q4 A0A147BIR5 A0A2M3ZI76 A0A2M3Z9F3 Q17D84 A0A2M4AP98 A0A182MZF0 A0A182P0G4 W5J506 A0A182VCS9 A0A182X1L7 Q7Q3D9 A0A182TSP1 A0A2S2R8C9 A0A0C9RJB1 A0A224XF19 A0A182F2Z5 A0A1Z5KWC0 A0A182L2F2 A0A182IEJ9 A0A2M4BRD8 A0A069DTD4 A0A182XZJ4 A0A1B6LVI1 A0A0V0GAL5 A0A182MNR4 A0A182RVK9 A0A1Y9IW62 A0A182K0B5 A0A084VBB7 A0A146LYP1 A0A0A9W8C8 A0A0K8SI43 J9JVZ1 A0A182JHW9 A0A1I8NB41 B0W195
A0A0L7LTI5 A0A0T6B8U5 A0A2A3E6L6 A0A087ZR85 A0A1L8DWE6 A0A1L8DWU5 E2BFY9 A0A2J7Q2E3 A0A067R9P4 A0A0L7QQK1 A0A195CUT9 A0A2J7Q2F7 F4WC33 E9IKB1 A0A0M9AAK4 A0A026WWD5 E2AH74 A0A154P5E5 A0A182PCI1 A0A232EPN5 A0A1B6CQ96 N6ULY0 A0A182M1I5 D6WH01 A0A195DLQ4 A0A195BKF4 A0A195FR16 A0A182WA52 A0A182QND2 A0A182YIM1 A0A182R7Q2 A0A1W4W9F7 A0A131XBC3 A0A158NAQ8 A0A310SJQ9 B0W1N3 Q7Q0U8 A0A182HQV4 A0A182UEW1 A0A182XLF4 A0A1S4J383 A0A2R5L7J8 A0A1Y1K5A4 A0A182NPW2 A0A182VB61 A0A0J7KSX3 A0A1Q3FJX7 A0A1Q3FJW6 L7LZP2 A0A2H8TRQ1 A0A182KJL0 A0A2J7Q2E5 A0A1J1J8A2 A0A084WGA6 B7PTK5 A0A131Y4D2 A0A182K4Q4 A0A147BIR5 A0A2M3ZI76 A0A2M3Z9F3 Q17D84 A0A2M4AP98 A0A182MZF0 A0A182P0G4 W5J506 A0A182VCS9 A0A182X1L7 Q7Q3D9 A0A182TSP1 A0A2S2R8C9 A0A0C9RJB1 A0A224XF19 A0A182F2Z5 A0A1Z5KWC0 A0A182L2F2 A0A182IEJ9 A0A2M4BRD8 A0A069DTD4 A0A182XZJ4 A0A1B6LVI1 A0A0V0GAL5 A0A182MNR4 A0A182RVK9 A0A1Y9IW62 A0A182K0B5 A0A084VBB7 A0A146LYP1 A0A0A9W8C8 A0A0K8SI43 J9JVZ1 A0A182JHW9 A0A1I8NB41 B0W195
EC Number
2.7.7.7
Pubmed
19121390
22118469
26354079
26227816
20798317
24845553
+ More
21719571 21282665 24508170 30249741 28648823 23537049 18362917 19820115 25244985 28049606 21347285 12364791 14747013 17210077 28004739 25576852 20966253 24438588 29652888 17510324 20920257 23761445 28528879 26334808 26823975 25401762 25315136
21719571 21282665 24508170 30249741 28648823 23537049 18362917 19820115 25244985 28049606 21347285 12364791 14747013 17210077 28004739 25576852 20966253 24438588 29652888 17510324 20920257 23761445 28528879 26334808 26823975 25401762 25315136
EMBL
BABH01034423
BABH01034424
BABH01044827
BABH01044828
BABH01044829
NWSH01000050
+ More
PCG80200.1 AGBW02013812 OWR42718.1 KQ458860 KPJ04756.1 KQ461191 KPJ07103.1 JTDY01000114 KOB78775.1 LJIG01009203 KRT83549.1 KZ288375 PBC26691.1 GFDF01003341 JAV10743.1 GFDF01003176 JAV10908.1 GL448096 EFN85391.1 NEVH01019371 PNF22752.1 KK852798 KDR16352.1 KQ414786 KOC60898.1 KQ977276 KYN04448.1 PNF22753.1 GL888070 EGI68013.1 GL763956 EFZ18992.1 KQ435699 KOX80587.1 KK107079 QOIP01000010 EZA60113.1 RLU17658.1 GL439483 EFN67191.1 KQ434820 KZC07071.1 NNAY01002921 OXU20292.1 GEDC01021611 JAS15687.1 APGK01026886 APGK01026887 KB740648 ENN79717.1 AXCM01000963 KQ971321 EFA00612.2 KQ980734 KYN13776.1 KQ976455 KYM85095.1 KQ981305 KYN42913.1 AXCN02001998 GEFH01005196 JAP63385.1 ADTU01009954 KQ765605 OAD53830.1 DS231823 EDS26584.1 AAAB01008980 EAA14002.4 APCN01001603 GGLE01001309 MBY05435.1 GEZM01092331 JAV56594.1 LBMM01003514 KMQ93438.1 GFDL01007221 JAV27824.1 GFDL01007231 JAV27814.1 GACK01007982 JAA57052.1 GFXV01005068 MBW16873.1 PNF22754.1 CVRI01000074 CRL07996.1 ATLV01023499 KE525344 KFB49250.1 ABJB010185757 ABJB010302451 ABJB010502063 ABJB010635012 ABJB010799269 ABJB011122057 DS786398 EEC09927.1 GEFM01002109 JAP73687.1 GEGO01005209 JAR90195.1 GGFM01007495 MBW28246.1 GGFM01004415 MBW25166.1 CH477298 EAT44345.1 GGFK01009279 MBW42600.1 ADMH02002186 ETN57925.1 AAAB01008964 EAA12253.4 GGMS01016419 MBY85622.1 GBYB01008295 JAG78062.1 GFTR01005351 JAW11075.1 GFJQ02007573 JAV99396.1 APCN01005204 GGFJ01006441 MBW55582.1 GBGD01001977 JAC86912.1 GEBQ01012299 JAT27678.1 GECL01001035 JAP05089.1 AXCM01006351 ATLV01006318 KE524352 KFB35261.1 GDHC01007169 JAQ11460.1 GBHO01038897 JAG04707.1 GBRD01013032 JAG52794.1 ABLF02010811 DS231821 EDS44965.1
PCG80200.1 AGBW02013812 OWR42718.1 KQ458860 KPJ04756.1 KQ461191 KPJ07103.1 JTDY01000114 KOB78775.1 LJIG01009203 KRT83549.1 KZ288375 PBC26691.1 GFDF01003341 JAV10743.1 GFDF01003176 JAV10908.1 GL448096 EFN85391.1 NEVH01019371 PNF22752.1 KK852798 KDR16352.1 KQ414786 KOC60898.1 KQ977276 KYN04448.1 PNF22753.1 GL888070 EGI68013.1 GL763956 EFZ18992.1 KQ435699 KOX80587.1 KK107079 QOIP01000010 EZA60113.1 RLU17658.1 GL439483 EFN67191.1 KQ434820 KZC07071.1 NNAY01002921 OXU20292.1 GEDC01021611 JAS15687.1 APGK01026886 APGK01026887 KB740648 ENN79717.1 AXCM01000963 KQ971321 EFA00612.2 KQ980734 KYN13776.1 KQ976455 KYM85095.1 KQ981305 KYN42913.1 AXCN02001998 GEFH01005196 JAP63385.1 ADTU01009954 KQ765605 OAD53830.1 DS231823 EDS26584.1 AAAB01008980 EAA14002.4 APCN01001603 GGLE01001309 MBY05435.1 GEZM01092331 JAV56594.1 LBMM01003514 KMQ93438.1 GFDL01007221 JAV27824.1 GFDL01007231 JAV27814.1 GACK01007982 JAA57052.1 GFXV01005068 MBW16873.1 PNF22754.1 CVRI01000074 CRL07996.1 ATLV01023499 KE525344 KFB49250.1 ABJB010185757 ABJB010302451 ABJB010502063 ABJB010635012 ABJB010799269 ABJB011122057 DS786398 EEC09927.1 GEFM01002109 JAP73687.1 GEGO01005209 JAR90195.1 GGFM01007495 MBW28246.1 GGFM01004415 MBW25166.1 CH477298 EAT44345.1 GGFK01009279 MBW42600.1 ADMH02002186 ETN57925.1 AAAB01008964 EAA12253.4 GGMS01016419 MBY85622.1 GBYB01008295 JAG78062.1 GFTR01005351 JAW11075.1 GFJQ02007573 JAV99396.1 APCN01005204 GGFJ01006441 MBW55582.1 GBGD01001977 JAC86912.1 GEBQ01012299 JAT27678.1 GECL01001035 JAP05089.1 AXCM01006351 ATLV01006318 KE524352 KFB35261.1 GDHC01007169 JAQ11460.1 GBHO01038897 JAG04707.1 GBRD01013032 JAG52794.1 ABLF02010811 DS231821 EDS44965.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000242457 UP000005203 UP000008237 UP000235965 UP000027135 UP000053825 UP000078542 UP000007755 UP000053105 UP000053097 UP000279307 UP000000311 UP000076502 UP000075885 UP000215335 UP000019118 UP000075883 UP000007266 UP000078492 UP000078540 UP000078541 UP000075920 UP000075886 UP000076408 UP000075900 UP000192223 UP000005205 UP000002320 UP000007062 UP000075840 UP000075902 UP000076407 UP000075884 UP000075903 UP000036403 UP000075882 UP000183832 UP000030765 UP000001555 UP000075881 UP000008820 UP000000673 UP000069272 UP000007819 UP000075880 UP000095301
UP000242457 UP000005203 UP000008237 UP000235965 UP000027135 UP000053825 UP000078542 UP000007755 UP000053105 UP000053097 UP000279307 UP000000311 UP000076502 UP000075885 UP000215335 UP000019118 UP000075883 UP000007266 UP000078492 UP000078540 UP000078541 UP000075920 UP000075886 UP000076408 UP000075900 UP000192223 UP000005205 UP000002320 UP000007062 UP000075840 UP000075902 UP000076407 UP000075884 UP000075903 UP000036403 UP000075882 UP000183832 UP000030765 UP000001555 UP000075881 UP000008820 UP000000673 UP000069272 UP000007819 UP000075880 UP000095301
Pfam
Interpro
IPR029052
Metallo-depent_PP-like
+ More
IPR033308 PGAP5/Cdc1/Ted1
IPR004843 Calcineurin-like_PHP_ApaH
IPR003583 Hlx-hairpin-Hlx_DNA-bd_motif
IPR019843 DNA_pol-X_BS
IPR018944 DNA_pol_lambd_fingers_domain
IPR037160 DNA_Pol_thumb_sf
IPR002008 DNA_pol_X_beta-like
IPR010996 DNA_pol_b-like_N
IPR027421 DNA_pol_lamdba_lyase_dom_sf
IPR002054 DNA-dir_DNA_pol_X
IPR022312 DNA_pol_X
IPR029398 PolB_thumb
IPR028207 DNA_pol_B_palm_palm
IPR033308 PGAP5/Cdc1/Ted1
IPR004843 Calcineurin-like_PHP_ApaH
IPR003583 Hlx-hairpin-Hlx_DNA-bd_motif
IPR019843 DNA_pol-X_BS
IPR018944 DNA_pol_lambd_fingers_domain
IPR037160 DNA_Pol_thumb_sf
IPR002008 DNA_pol_X_beta-like
IPR010996 DNA_pol_b-like_N
IPR027421 DNA_pol_lamdba_lyase_dom_sf
IPR002054 DNA-dir_DNA_pol_X
IPR022312 DNA_pol_X
IPR029398 PolB_thumb
IPR028207 DNA_pol_B_palm_palm
SUPFAM
SSF47802
SSF47802
Gene 3D
CDD
ProteinModelPortal
H9J2P1
H9JXP7
A0A2A4K919
A0A212EMK8
A0A194QMP4
A0A194QNK3
+ More
A0A0L7LTI5 A0A0T6B8U5 A0A2A3E6L6 A0A087ZR85 A0A1L8DWE6 A0A1L8DWU5 E2BFY9 A0A2J7Q2E3 A0A067R9P4 A0A0L7QQK1 A0A195CUT9 A0A2J7Q2F7 F4WC33 E9IKB1 A0A0M9AAK4 A0A026WWD5 E2AH74 A0A154P5E5 A0A182PCI1 A0A232EPN5 A0A1B6CQ96 N6ULY0 A0A182M1I5 D6WH01 A0A195DLQ4 A0A195BKF4 A0A195FR16 A0A182WA52 A0A182QND2 A0A182YIM1 A0A182R7Q2 A0A1W4W9F7 A0A131XBC3 A0A158NAQ8 A0A310SJQ9 B0W1N3 Q7Q0U8 A0A182HQV4 A0A182UEW1 A0A182XLF4 A0A1S4J383 A0A2R5L7J8 A0A1Y1K5A4 A0A182NPW2 A0A182VB61 A0A0J7KSX3 A0A1Q3FJX7 A0A1Q3FJW6 L7LZP2 A0A2H8TRQ1 A0A182KJL0 A0A2J7Q2E5 A0A1J1J8A2 A0A084WGA6 B7PTK5 A0A131Y4D2 A0A182K4Q4 A0A147BIR5 A0A2M3ZI76 A0A2M3Z9F3 Q17D84 A0A2M4AP98 A0A182MZF0 A0A182P0G4 W5J506 A0A182VCS9 A0A182X1L7 Q7Q3D9 A0A182TSP1 A0A2S2R8C9 A0A0C9RJB1 A0A224XF19 A0A182F2Z5 A0A1Z5KWC0 A0A182L2F2 A0A182IEJ9 A0A2M4BRD8 A0A069DTD4 A0A182XZJ4 A0A1B6LVI1 A0A0V0GAL5 A0A182MNR4 A0A182RVK9 A0A1Y9IW62 A0A182K0B5 A0A084VBB7 A0A146LYP1 A0A0A9W8C8 A0A0K8SI43 J9JVZ1 A0A182JHW9 A0A1I8NB41 B0W195
A0A0L7LTI5 A0A0T6B8U5 A0A2A3E6L6 A0A087ZR85 A0A1L8DWE6 A0A1L8DWU5 E2BFY9 A0A2J7Q2E3 A0A067R9P4 A0A0L7QQK1 A0A195CUT9 A0A2J7Q2F7 F4WC33 E9IKB1 A0A0M9AAK4 A0A026WWD5 E2AH74 A0A154P5E5 A0A182PCI1 A0A232EPN5 A0A1B6CQ96 N6ULY0 A0A182M1I5 D6WH01 A0A195DLQ4 A0A195BKF4 A0A195FR16 A0A182WA52 A0A182QND2 A0A182YIM1 A0A182R7Q2 A0A1W4W9F7 A0A131XBC3 A0A158NAQ8 A0A310SJQ9 B0W1N3 Q7Q0U8 A0A182HQV4 A0A182UEW1 A0A182XLF4 A0A1S4J383 A0A2R5L7J8 A0A1Y1K5A4 A0A182NPW2 A0A182VB61 A0A0J7KSX3 A0A1Q3FJX7 A0A1Q3FJW6 L7LZP2 A0A2H8TRQ1 A0A182KJL0 A0A2J7Q2E5 A0A1J1J8A2 A0A084WGA6 B7PTK5 A0A131Y4D2 A0A182K4Q4 A0A147BIR5 A0A2M3ZI76 A0A2M3Z9F3 Q17D84 A0A2M4AP98 A0A182MZF0 A0A182P0G4 W5J506 A0A182VCS9 A0A182X1L7 Q7Q3D9 A0A182TSP1 A0A2S2R8C9 A0A0C9RJB1 A0A224XF19 A0A182F2Z5 A0A1Z5KWC0 A0A182L2F2 A0A182IEJ9 A0A2M4BRD8 A0A069DTD4 A0A182XZJ4 A0A1B6LVI1 A0A0V0GAL5 A0A182MNR4 A0A182RVK9 A0A1Y9IW62 A0A182K0B5 A0A084VBB7 A0A146LYP1 A0A0A9W8C8 A0A0K8SI43 J9JVZ1 A0A182JHW9 A0A1I8NB41 B0W195
Ontologies
GO
PANTHER
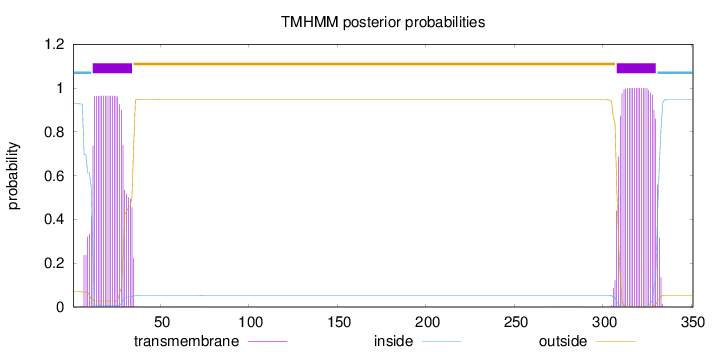
Topology
Subcellular location
Nucleus
Length:
351
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.9917099999999
Exp number, first 60 AAs:
21.07887
Total prob of N-in:
0.92813
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 307
TMhelix
308 - 330
inside
331 - 351
Population Genetic Test Statistics
Pi
139.397444
Theta
136.843833
Tajima's D
-0.340213
CLR
1.803083
CSRT
0.274386280685966
Interpretation
Uncertain
