Gene
KWMTBOMO02295
Pre Gene Modal
BGIBMGA003777
Annotation
PREDICTED:_cyclin_3_isoform_X2_[Bombyx_mori]
Full name
G2/mitotic-specific cyclin-B3
Location in the cell
Mitochondrial Reliability : 2.651
Sequence
CDS
ATGGCGCCAACAAGATTGGCAACGAAACCGCTCGGCGTCAACGAGAACCTGAACGGCATGTTAAGCCACGGCATTACGACGAGGAGTAAACAGCTAGCTTCTCTAAAGAGTCACAAGGACAGTCTAAAAATTCCCAATAAAAGGAAAGCGGACAGTCCTCTTAAAGATGTGACTGCGAAGAGGGCGGCGTTCGGAGATATAACGAACGCCTATAACAAATCGTGCGCGCCCCAGGACAAGGATAAAGCCATGCCAGTGAAGAAAGTCACTGTTGAGACCAAAATGCTTCCAGCAGTAAAGAGCATCAAATCACAAACAAGCACACTAAATGGCAAAACTTCAAAGACCAAACAACTCCAGCGGGTAGCCAATAATGCTATTGATGCAGTACAGAAACATTTCACCAAAGACCCACCCAAGCAAGTGGCCACAAGAGCTCAGAATGGCATCCTGAAGAATTTTGAAAGGAAAGACAAACTCAAAGAGCAGAATCTGAGTAATGGTTCAGTCAAATCTGCAAGAAGTGATGTTTCCGGTGAACCCTCACTTTATATGACTGCCTTGGAGAATATCCCATCTGAGGTAACTAAAGAAGAGTACAAACAAATCAGTGAGAAACTGAACAAAGTAAACTTGGATGACACATCTAGGTCTTTGGATGAAGAAACAGAGACCCCTCATAAAACTCCTTCTGGTGTTCTTGACTTTGACTTTGAGAATTGGAATGATCCACTCCAAGTATCTCATTATGCTATGGACATATTTAATTATTTGAAAAACAGGGAGCGCCTTTTCCCTATTGACGATTACCTGAAACGTATGAATGGAATTACATCGTGGATGAGAGCGCTACTTGTAGACTGGATGGTTGAAGTTCAAGAGAGTTTTGAACTCAATCATGAGACCCTGTATTTGGCTGTGAAGCTAGTTGACTTGTTCCTTACTCGTTCAACTAAGAATGAACAAATAGAACAACTGACAAAGGAGGAATTGCAACTATTAGGAGCATCTGCCTTGTTTATAGCTTCTAAGTTTGATGAGCGCATTCCGCCCCTAGTGGACGACTTCCTGTACATCTGCGACGGGGCATATACGCTGAGCCAACTGCTTAAAATGGAGATGACTATTCTCCGGGTCATCGACTTCGATCTCGGCGTGCCGCTCTCCTACCGGTTCCTCAGGAGATACGCCCGGTGTGCTCGTGTATCAATGCCTACACTAACACTGGCTCGGTTTGTGTTAGAACAATGCTTGTTGGAATATGGACTTATTGAAAATTCAGACAGTAAAATGGCTGCTGCTGCACTGTACCTTGCTCTCAGGATGAAGAATCTAGGTTCATGGACACCCACTCTACAGTATTATACAGGTTACAACCTCAAAGAAATTCTCCCGATTGCATTAGCCCAGAATGCAGTTCTACACAAGAAGCCAAAGTCTGCGATTAGCACAGTTAGGAATAAATACTCACATACAATATTTTTCGAAGTTGCCAAAGTACCCCTAATAGACGAGGCGACGCTCACTAGCTGA
Protein
MAPTRLATKPLGVNENLNGMLSHGITTRSKQLASLKSHKDSLKIPNKRKADSPLKDVTAKRAAFGDITNAYNKSCAPQDKDKAMPVKKVTVETKMLPAVKSIKSQTSTLNGKTSKTKQLQRVANNAIDAVQKHFTKDPPKQVATRAQNGILKNFERKDKLKEQNLSNGSVKSARSDVSGEPSLYMTALENIPSEVTKEEYKQISEKLNKVNLDDTSRSLDEETETPHKTPSGVLDFDFENWNDPLQVSHYAMDIFNYLKNRERLFPIDDYLKRMNGITSWMRALLVDWMVEVQESFELNHETLYLAVKLVDLFLTRSTKNEQIEQLTKEELQLLGASALFIASKFDERIPPLVDDFLYICDGAYTLSQLLKMEMTILRVIDFDLGVPLSYRFLRRYARCARVSMPTLTLARFVLEQCLLEYGLIENSDSKMAAAALYLALRMKNLGSWTPTLQYYTGYNLKEILPIALAQNAVLHKKPKSAISTVRNKYSHTIFFEVAKVPLIDEATLTS
Summary
Description
Cyclins are positive regulatory subunits of the cyclin-dependent kinases (CDKs), and thereby play an essential role in the control of the cell cycle, notably via their destruction during cell division. Probably functions redundantly with other cyclins in regulation of cell cycle. Its presence may be required to delay a deadline for completing cytokinesis that is ordinary imposed by nuclear envelope reformation. Degradation of CycB and CycB3 promote cytokinesis furrow initiation and ingression. Required with CycB for female fertility.
Subunit
Interacts with Cdk1 kinase.
Similarity
Belongs to the cyclin family.
Belongs to the cyclin family. Cyclin AB subfamily.
Belongs to the cyclin family. Cyclin AB subfamily.
Keywords
Cell cycle
Cell division
Complete proteome
Cyclin
Mitosis
Nucleus
Phosphoprotein
Reference proteome
Ubl conjugation
Feature
chain G2/mitotic-specific cyclin-B3
Uniprot
H9J2P0
A8HI21
A0A2A4K8E2
A0A2H1W667
S4P8S9
A0A194QH62
+ More
A0A212EML7 A0A0L7LJJ6 A0A194QQ62 K7J596 A0A291S6V0 A0A232FGF7 A0A154PNJ3 A0A0M9A5V3 A0A1W4WNA3 A0A0L7QNT0 A0A088A1H8 A0A2A3EJA1 A0A2J7R661 E2C2R2 A0A067QUB1 A0A195CE28 A0A195BKA8 A0A0J7KUF7 D6WNC8 F4X2P3 E9J286 A0A1Y1L7J4 A0A151WNA0 A0A195DSL3 A0A1A9WYR9 A0A1A9UVG4 E0VTL6 A0A0K8UTM8 Q29CG1 E1ZXN0 A0A034VGF1 B3LV01 B4LWX7 A0A0A1WNS0 A0A0B4KHT9 Q9I7I0 A0A1I8PBZ4 B0X498 B4QTU7 A0A3B0JHH9 A0A336MP55 B4PV23 B4GN38 B4K8C3 B3P6W4 A0A1W4W2G5 B4NG21 A0A0M3QYL5 A0A1Q3EWM2 A0A1Q3EWM8 A0A1B0CAS6 A0A1L8DWW8 W8CDF8 A0A1J1IIC6 B4HHM7 A0A1S4FMV3 Q16VG6 A0A0P4WP59 A0A0P4WGL0 A0A1B6G928 A0A2P2I6B5 N6U2D3 A0A023EZ14 A0A1S3DRG2
A0A212EML7 A0A0L7LJJ6 A0A194QQ62 K7J596 A0A291S6V0 A0A232FGF7 A0A154PNJ3 A0A0M9A5V3 A0A1W4WNA3 A0A0L7QNT0 A0A088A1H8 A0A2A3EJA1 A0A2J7R661 E2C2R2 A0A067QUB1 A0A195CE28 A0A195BKA8 A0A0J7KUF7 D6WNC8 F4X2P3 E9J286 A0A1Y1L7J4 A0A151WNA0 A0A195DSL3 A0A1A9WYR9 A0A1A9UVG4 E0VTL6 A0A0K8UTM8 Q29CG1 E1ZXN0 A0A034VGF1 B3LV01 B4LWX7 A0A0A1WNS0 A0A0B4KHT9 Q9I7I0 A0A1I8PBZ4 B0X498 B4QTU7 A0A3B0JHH9 A0A336MP55 B4PV23 B4GN38 B4K8C3 B3P6W4 A0A1W4W2G5 B4NG21 A0A0M3QYL5 A0A1Q3EWM2 A0A1Q3EWM8 A0A1B0CAS6 A0A1L8DWW8 W8CDF8 A0A1J1IIC6 B4HHM7 A0A1S4FMV3 Q16VG6 A0A0P4WP59 A0A0P4WGL0 A0A1B6G928 A0A2P2I6B5 N6U2D3 A0A023EZ14 A0A1S3DRG2
Pubmed
19121390
23622113
26354079
22118469
26227816
20075255
+ More
28992199 28648823 20798317 24845553 18362917 19820115 21719571 21282665 28004739 20566863 15632085 17994087 25348373 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9851980 7588612 11369230 12620185 18327897 17550304 24495485 17510324 23537049 25474469
28992199 28648823 20798317 24845553 18362917 19820115 21719571 21282665 28004739 20566863 15632085 17994087 25348373 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9851980 7588612 11369230 12620185 18327897 17550304 24495485 17510324 23537049 25474469
EMBL
BABH01034420
EU074796
ABV66133.1
NWSH01000050
PCG80198.1
ODYU01006605
+ More
SOQ48595.1 GAIX01004059 JAA88501.1 KQ458860 KPJ04754.1 AGBW02013812 OWR42716.1 JTDY01000890 KOB75539.1 KQ461191 KPJ07105.1 AAZX01000332 MF432988 ATL75339.1 NNAY01000239 OXU29782.1 KQ434978 KZC13034.1 KQ435748 KOX76479.1 KQ414851 KOC60224.1 KZ288241 PBC31269.1 NEVH01006980 PNF36324.1 GL452203 EFN77747.1 KK853004 KDR12607.1 KQ977957 KYM98486.1 KQ976450 KYM85705.1 LBMM01003109 KMQ93953.1 KQ971343 EFA03235.1 GL888591 EGI59239.1 GL767834 EFZ13064.1 GEZM01066382 JAV67915.1 KQ982919 KYQ49271.1 KQ980489 KYN15838.1 DS235767 EEB16691.1 GDHF01022451 JAI29863.1 CM000070 EAL26685.1 GL435067 EFN74068.1 GAKP01016566 JAC42386.1 CH902617 EDV42473.1 CH940650 EDW67724.1 GBXI01013800 JAD00492.1 AE014297 AGB96314.1 AJ006772 AJ012568 BT003185 DS232333 EDS40251.1 CM000364 EDX14301.1 OUUW01000005 SPP80163.1 UFQT01001658 SSX31331.1 CM000160 EDW98872.1 CH479186 EDW39164.1 CH933806 EDW16505.1 CH954182 EDV53784.1 CH964251 EDW83238.1 CP012526 ALC47829.1 GFDL01015328 JAV19717.1 GFDL01015326 JAV19719.1 AJWK01004228 GFDF01003168 JAV10916.1 GAMC01002021 JAC04535.1 CVRI01000054 CRK99997.1 CH480815 EDW43569.1 CH477593 EAT38552.1 GDRN01055273 JAI65998.1 GDRN01055272 JAI65999.1 GECZ01010850 JAS58919.1 IACF01003962 LAB69565.1 APGK01044620 KB741028 ENN74781.1 GBBI01004267 JAC14445.1
SOQ48595.1 GAIX01004059 JAA88501.1 KQ458860 KPJ04754.1 AGBW02013812 OWR42716.1 JTDY01000890 KOB75539.1 KQ461191 KPJ07105.1 AAZX01000332 MF432988 ATL75339.1 NNAY01000239 OXU29782.1 KQ434978 KZC13034.1 KQ435748 KOX76479.1 KQ414851 KOC60224.1 KZ288241 PBC31269.1 NEVH01006980 PNF36324.1 GL452203 EFN77747.1 KK853004 KDR12607.1 KQ977957 KYM98486.1 KQ976450 KYM85705.1 LBMM01003109 KMQ93953.1 KQ971343 EFA03235.1 GL888591 EGI59239.1 GL767834 EFZ13064.1 GEZM01066382 JAV67915.1 KQ982919 KYQ49271.1 KQ980489 KYN15838.1 DS235767 EEB16691.1 GDHF01022451 JAI29863.1 CM000070 EAL26685.1 GL435067 EFN74068.1 GAKP01016566 JAC42386.1 CH902617 EDV42473.1 CH940650 EDW67724.1 GBXI01013800 JAD00492.1 AE014297 AGB96314.1 AJ006772 AJ012568 BT003185 DS232333 EDS40251.1 CM000364 EDX14301.1 OUUW01000005 SPP80163.1 UFQT01001658 SSX31331.1 CM000160 EDW98872.1 CH479186 EDW39164.1 CH933806 EDW16505.1 CH954182 EDV53784.1 CH964251 EDW83238.1 CP012526 ALC47829.1 GFDL01015328 JAV19717.1 GFDL01015326 JAV19719.1 AJWK01004228 GFDF01003168 JAV10916.1 GAMC01002021 JAC04535.1 CVRI01000054 CRK99997.1 CH480815 EDW43569.1 CH477593 EAT38552.1 GDRN01055273 JAI65998.1 GDRN01055272 JAI65999.1 GECZ01010850 JAS58919.1 IACF01003962 LAB69565.1 APGK01044620 KB741028 ENN74781.1 GBBI01004267 JAC14445.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000053240
+ More
UP000002358 UP000215335 UP000076502 UP000053105 UP000192223 UP000053825 UP000005203 UP000242457 UP000235965 UP000008237 UP000027135 UP000078542 UP000078540 UP000036403 UP000007266 UP000007755 UP000075809 UP000078492 UP000091820 UP000078200 UP000009046 UP000001819 UP000000311 UP000007801 UP000008792 UP000000803 UP000095300 UP000002320 UP000000304 UP000268350 UP000002282 UP000008744 UP000009192 UP000008711 UP000192221 UP000007798 UP000092553 UP000092461 UP000183832 UP000001292 UP000008820 UP000019118 UP000079169
UP000002358 UP000215335 UP000076502 UP000053105 UP000192223 UP000053825 UP000005203 UP000242457 UP000235965 UP000008237 UP000027135 UP000078542 UP000078540 UP000036403 UP000007266 UP000007755 UP000075809 UP000078492 UP000091820 UP000078200 UP000009046 UP000001819 UP000000311 UP000007801 UP000008792 UP000000803 UP000095300 UP000002320 UP000000304 UP000268350 UP000002282 UP000008744 UP000009192 UP000008711 UP000192221 UP000007798 UP000092553 UP000092461 UP000183832 UP000001292 UP000008820 UP000019118 UP000079169
Interpro
SUPFAM
SSF47954
SSF47954
CDD
ProteinModelPortal
H9J2P0
A8HI21
A0A2A4K8E2
A0A2H1W667
S4P8S9
A0A194QH62
+ More
A0A212EML7 A0A0L7LJJ6 A0A194QQ62 K7J596 A0A291S6V0 A0A232FGF7 A0A154PNJ3 A0A0M9A5V3 A0A1W4WNA3 A0A0L7QNT0 A0A088A1H8 A0A2A3EJA1 A0A2J7R661 E2C2R2 A0A067QUB1 A0A195CE28 A0A195BKA8 A0A0J7KUF7 D6WNC8 F4X2P3 E9J286 A0A1Y1L7J4 A0A151WNA0 A0A195DSL3 A0A1A9WYR9 A0A1A9UVG4 E0VTL6 A0A0K8UTM8 Q29CG1 E1ZXN0 A0A034VGF1 B3LV01 B4LWX7 A0A0A1WNS0 A0A0B4KHT9 Q9I7I0 A0A1I8PBZ4 B0X498 B4QTU7 A0A3B0JHH9 A0A336MP55 B4PV23 B4GN38 B4K8C3 B3P6W4 A0A1W4W2G5 B4NG21 A0A0M3QYL5 A0A1Q3EWM2 A0A1Q3EWM8 A0A1B0CAS6 A0A1L8DWW8 W8CDF8 A0A1J1IIC6 B4HHM7 A0A1S4FMV3 Q16VG6 A0A0P4WP59 A0A0P4WGL0 A0A1B6G928 A0A2P2I6B5 N6U2D3 A0A023EZ14 A0A1S3DRG2
A0A212EML7 A0A0L7LJJ6 A0A194QQ62 K7J596 A0A291S6V0 A0A232FGF7 A0A154PNJ3 A0A0M9A5V3 A0A1W4WNA3 A0A0L7QNT0 A0A088A1H8 A0A2A3EJA1 A0A2J7R661 E2C2R2 A0A067QUB1 A0A195CE28 A0A195BKA8 A0A0J7KUF7 D6WNC8 F4X2P3 E9J286 A0A1Y1L7J4 A0A151WNA0 A0A195DSL3 A0A1A9WYR9 A0A1A9UVG4 E0VTL6 A0A0K8UTM8 Q29CG1 E1ZXN0 A0A034VGF1 B3LV01 B4LWX7 A0A0A1WNS0 A0A0B4KHT9 Q9I7I0 A0A1I8PBZ4 B0X498 B4QTU7 A0A3B0JHH9 A0A336MP55 B4PV23 B4GN38 B4K8C3 B3P6W4 A0A1W4W2G5 B4NG21 A0A0M3QYL5 A0A1Q3EWM2 A0A1Q3EWM8 A0A1B0CAS6 A0A1L8DWW8 W8CDF8 A0A1J1IIC6 B4HHM7 A0A1S4FMV3 Q16VG6 A0A0P4WP59 A0A0P4WGL0 A0A1B6G928 A0A2P2I6B5 N6U2D3 A0A023EZ14 A0A1S3DRG2
PDB
6GU4
E-value=1.94837e-41,
Score=426
Ontologies
GO
PANTHER
Topology
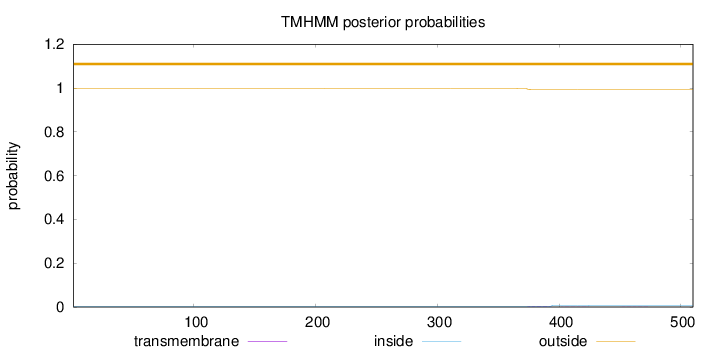
Subcellular location
Nucleus
Length:
510
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08833
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.00241
outside
1 - 510
Population Genetic Test Statistics
Pi
318.438694
Theta
216.498144
Tajima's D
1.263953
CLR
0
CSRT
0.733863306834658
Interpretation
Uncertain
