Pre Gene Modal
BGIBMGA003775
Annotation
PREDICTED:_cell_division_control_protein_48_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.614 Nuclear Reliability : 2.283
Sequence
CDS
ATGTTAGGAGGTGCTAAACTAAAAGTAGTGTTTACTCACGAAAAAACTTGCTTCGCATATATAAGTCCAAAGTACAGTAGTAATAATGAACAGTCTCAGTGCCTTCAAGTTCTATACAAAGACAAGCACCTGTTTTTATGGGTGGTGTACAGCAGTGGTGTTGCGGAAGGACAAATAGCTTGCAATCCAGTGCATAGTCAACTGATTGGCTTAGATGAGGGAGCTGATGTGTTTGTCTCACCATACGGTGATGTGCGAGTGCTGGATGAATTGTATATAGATACGGACAGCCCTGATGACCAGGAGTTGTTGGAACACAATGCGGAATTACTGCAGCTACGAATACTAGATCAGCTTCGATATGTGGCGGCACATCAAAAATGCGTCGTGTGGATATTTTCATCGATGCCCATCGTTTTCACGCCGAAGCAGACCGGCTTGCTAGTGAACCAGAGCCGCGTAGTCGTTAAAGTTAACGCGTTCAATAGCTACCAAGGCTTCCAAACAAAACCTCCAGCGCCAAACGAACCCAATGGCCCTTTTAAAAATATAGGCTTAATAAACGAAGAGATGCTTCGTCCATATTTGAACTCACCAAAGAGGCTTGTATTGCGTTCGGTACCTATAGACAGCGACGGCAAAAAGAATTTGATACATCCTTACACGGTTTTCATACATGAAGATTTAATAGACGAATGCTTCAAAAATTTAACTGTTATATTATCGACCATGAATTCTATTCCTTCGATTCTGAGCGAAACTGATGAAGACGATAAAGAGAACAATAATAACGCCATAAACGATTTGTGCGTTGAAATTGTGCCCATAGACGGTATCATATACAGGAGCTTGAGCCGCGAAGTGTACAACAGAAACATACCGACAGTGTTGATACCGAAATCGTTGAATGCCATTATCAACATTGAGAATGGAACGAGAATTATATTCAACATAATCGGGGATACGGTAGAGCAACCTGTGCACATTGAATTGGTGACGTATACAGAAGAAAAACAAACAGAAGTCGATGTTATAGACAGATTCAAGAAATGCGTCATAGAGAACACGCATTCAGGCAAAAGATTTTTAATAAATAACGGCATAGTTAAGCAGAATAGTCAGATAAGCGATGGTTATTTGCGGTTCAATTTGAAACCAGACGGTTTGAAATATACCCTCTTGGATTCGGAGAGTTTGAGGCATTTCACTACGAAGGCAACGTGCCTTAACGACACGGATTTAGTCCTGCCGAAACGCAGCGCCTCCCGCATGGAGTATGATTTCAAGAACTATTGCAGGACATTGAAATCAATAGAGACTTTGATCAGTAAAGTTCTCTCGCATTTGTATTTCGAAATACATAGAGAGGCGACGTTTAAAAATGCATCAGAGATTAAAAGCAACGTTCTGATCACGGGGTCCAGTGGTATGGGTAAAACATCGTTGTGTCACATAATCGAGAAGGAGCTGACGGTCTGGTCTCATACGTTGCACTGCAGGGCGCTCAGGGGCCGTAAGGATATACCGGAGATCCTGGGCAAAGCTATCCTCATGTGCCAAGAGCACAGTCCGGCTGTATTGATCTGCGACGACGTGGACTCCATGGTACCGCCCGATATGGAGGGAGTTTCGCCCCAGGATATAGCGTATTATCAGAGATTAGCCGTGGTCATCAAGCACCTCCTGCAGACGTGCGCGGGTGTGTGCGTGCTGATGACGTCACTCTCAATGAAGTCCCTGCACCCGGTCCTCAGGCAGTTCAACGGGAAGCCGCTGTTCACGGCGCACTTCGATATACCTGAAGTTGAACAGGATGAAAGAGTGGAGCTGTTCAAGCATTTGATGAACGAGAAGATCCGTCAGCAGTTCGAGGTAGCGGACGATGACGTCATCACGTTGGCCATGGACACCGCCTCGTGCACAGTGCGGGACATCATCGACCACTTCAATAAGAAGATATTTAAAGCCGTCAAGATGAAAAAAGCGAACCCAGACATTGCGAAGCCTCGTTTAGTCGAGGAAACTGCTAAGGACACGGAGAAAACTACCGCGTTCGACATATGGGGGCCGGTGGGGGGCTTGGAGGACGTCAAGCGGGAACTCACGGAGTGTATCTTCTGGCCTGTCATGTACCCAGCGTTGTTCCCGTCACAATCCTGTGGGATCCTACTGTACGGTCCCCCCGGAACCGGCAAGTCCTACATCGGGTCCTGTCTCGCCAGACTCAACAATATGCACATGATAACAGTGAAAGGACCAGAGCTTCTCTCCAAGTACATAGGGCAGAGCGAGAAGGCTGTTCGGGATGTCTTCGACAAAGCGGACATGAAGCGTCCTTGCATATTGTTCTTCGACGAATTCGACAGTCTCGCGCCCAAACGTGGTCACGACTCGACCGGGGTGACGGACAGAGTGGTGAACCAGCTGCTGGCGAGGATGGACGGCGCCGAGGGCGGGGCCAGGGGTCCCGTTATAGCGGCCACATCGAGGCCGGACCTGGTGGACCCGGCGCTGCTGAGAGCCGGCAGACTTCAGAAGCATATCTACTGCGAACTGCCCGATAGGTGGGGCAGACACGAAGTACTCCAAACATTGACGAAGAATCTAACCGTAGACCAAGAGGTAGACCTCCGAGAGTTGTCTGAGCGCACCGAGGGCTACACTCCCGCCGATCTCAAGTCCTTACTGGTCACGGCGCAACTGACAAGACTAGAGAAACAATTGGCTCACAACGACGACACGACATTGGGGTCCGTGGTGGTGCAGCAAGAAGACATCGATGGAGCTTTGGAGGAAACCAAACCTTCGCTGTCGAGAGAACAATTACTGTTCTACGATATGATATACAAAAGGTTCAGAGGCGAGACTCTGACGTCCGAACAGAAGATAATGGCCGGCCGGTTTGAGAAGCAACGCGCCACTTTGGCTTAA
Protein
MLGGAKLKVVFTHEKTCFAYISPKYSSNNEQSQCLQVLYKDKHLFLWVVYSSGVAEGQIACNPVHSQLIGLDEGADVFVSPYGDVRVLDELYIDTDSPDDQELLEHNAELLQLRILDQLRYVAAHQKCVVWIFSSMPIVFTPKQTGLLVNQSRVVVKVNAFNSYQGFQTKPPAPNEPNGPFKNIGLINEEMLRPYLNSPKRLVLRSVPIDSDGKKNLIHPYTVFIHEDLIDECFKNLTVILSTMNSIPSILSETDEDDKENNNNAINDLCVEIVPIDGIIYRSLSREVYNRNIPTVLIPKSLNAIINIENGTRIIFNIIGDTVEQPVHIELVTYTEEKQTEVDVIDRFKKCVIENTHSGKRFLINNGIVKQNSQISDGYLRFNLKPDGLKYTLLDSESLRHFTTKATCLNDTDLVLPKRSASRMEYDFKNYCRTLKSIETLISKVLSHLYFEIHREATFKNASEIKSNVLITGSSGMGKTSLCHIIEKELTVWSHTLHCRALRGRKDIPEILGKAILMCQEHSPAVLICDDVDSMVPPDMEGVSPQDIAYYQRLAVVIKHLLQTCAGVCVLMTSLSMKSLHPVLRQFNGKPLFTAHFDIPEVEQDERVELFKHLMNEKIRQQFEVADDDVITLAMDTASCTVRDIIDHFNKKIFKAVKMKKANPDIAKPRLVEETAKDTEKTTAFDIWGPVGGLEDVKRELTECIFWPVMYPALFPSQSCGILLYGPPGTGKSYIGSCLARLNNMHMITVKGPELLSKYIGQSEKAVRDVFDKADMKRPCILFFDEFDSLAPKRGHDSTGVTDRVVNQLLARMDGAEGGARGPVIAATSRPDLVDPALLRAGRLQKHIYCELPDRWGRHEVLQTLTKNLTVDQEVDLRELSERTEGYTPADLKSLLVTAQLTRLEKQLAHNDDTTLGSVVVQQEDIDGALEETKPSLSREQLLFYDMIYKRFRGETLTSEQKIMAGRFEKQRATLA
Summary
Uniprot
A0A2A4K7I3
A0A2H1W6M7
A0A194QGT5
A0A194QU80
A0A212EMK0
A0A088A1K8
+ More
A0A0L7QNM3 D6WNC9 A0A182K513 B4IYS0 A0A0Q9WVR9 A0A232FGG9 A0A1Q3FC69 A0A1Q3FCM6 K7J597 A0A0Q9XMU1 B0XEN9 A0A0Q9XRI0 A0A034WFN0 A0A034WD20 A0A0K8US19 A0A3B0JL47 B4MKJ9 A0A0V0G6S1 B5DPB6 A0A0A9WLA2 B4GRZ9 A0A1W4U6U0 B4QJK5 Q17MW1 Q9Y090 Q9VUC7 A0A182HCR9 B3ND11 B4HHC1 B3M3I2 B4PI00 A0A151WN25
A0A0L7QNM3 D6WNC9 A0A182K513 B4IYS0 A0A0Q9WVR9 A0A232FGG9 A0A1Q3FC69 A0A1Q3FCM6 K7J597 A0A0Q9XMU1 B0XEN9 A0A0Q9XRI0 A0A034WFN0 A0A034WD20 A0A0K8US19 A0A3B0JL47 B4MKJ9 A0A0V0G6S1 B5DPB6 A0A0A9WLA2 B4GRZ9 A0A1W4U6U0 B4QJK5 Q17MW1 Q9Y090 Q9VUC7 A0A182HCR9 B3ND11 B4HHC1 B3M3I2 B4PI00 A0A151WN25
Pubmed
EMBL
NWSH01000050
PCG80197.1
ODYU01006605
SOQ48596.1
KQ458860
KPJ04753.1
+ More
KQ461191 KPJ07106.1 AGBW02013812 OWR42715.1 KQ414851 KOC60225.1 KQ971343 EFA04352.1 CH916366 EDV96607.1 CH940647 KRF84891.1 NNAY01000239 OXU29785.1 GFDL01009870 JAV25175.1 GFDL01009745 JAV25300.1 AAZX01000332 CH933809 KRG06628.1 DS232848 EDS26100.1 KRG06627.1 GAKP01006374 JAC52578.1 GAKP01006373 JAC52579.1 GDHF01023036 JAI29278.1 OUUW01000002 SPP76100.1 CH963847 EDW72705.2 GECL01002314 JAP03810.1 CH379069 EDY73556.2 GBHO01036306 JAG07298.1 CH479188 EDW40534.1 CM000363 CM002912 EDX10414.1 KMY99524.1 CH477203 EAT48041.1 AJ243811 CAB51031.1 AE014296 AY075423 AAF49760.1 AAL68239.1 JXUM01034490 KQ561024 KXJ79987.1 CH954178 EDV51733.1 CH480815 EDW41446.1 CH902618 EDV39243.2 CM000159 EDW94475.1 KQ982919 KYQ49274.1
KQ461191 KPJ07106.1 AGBW02013812 OWR42715.1 KQ414851 KOC60225.1 KQ971343 EFA04352.1 CH916366 EDV96607.1 CH940647 KRF84891.1 NNAY01000239 OXU29785.1 GFDL01009870 JAV25175.1 GFDL01009745 JAV25300.1 AAZX01000332 CH933809 KRG06628.1 DS232848 EDS26100.1 KRG06627.1 GAKP01006374 JAC52578.1 GAKP01006373 JAC52579.1 GDHF01023036 JAI29278.1 OUUW01000002 SPP76100.1 CH963847 EDW72705.2 GECL01002314 JAP03810.1 CH379069 EDY73556.2 GBHO01036306 JAG07298.1 CH479188 EDW40534.1 CM000363 CM002912 EDX10414.1 KMY99524.1 CH477203 EAT48041.1 AJ243811 CAB51031.1 AE014296 AY075423 AAF49760.1 AAL68239.1 JXUM01034490 KQ561024 KXJ79987.1 CH954178 EDV51733.1 CH480815 EDW41446.1 CH902618 EDV39243.2 CM000159 EDW94475.1 KQ982919 KYQ49274.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000005203
UP000053825
+ More
UP000007266 UP000075881 UP000001070 UP000008792 UP000215335 UP000002358 UP000009192 UP000002320 UP000268350 UP000007798 UP000001819 UP000008744 UP000192221 UP000000304 UP000008820 UP000000803 UP000069940 UP000249989 UP000008711 UP000001292 UP000007801 UP000002282 UP000075809
UP000007266 UP000075881 UP000001070 UP000008792 UP000215335 UP000002358 UP000009192 UP000002320 UP000268350 UP000007798 UP000001819 UP000008744 UP000192221 UP000000304 UP000008820 UP000000803 UP000069940 UP000249989 UP000008711 UP000001292 UP000007801 UP000002282 UP000075809
PRIDE
Interpro
ProteinModelPortal
A0A2A4K7I3
A0A2H1W6M7
A0A194QGT5
A0A194QU80
A0A212EMK0
A0A088A1K8
+ More
A0A0L7QNM3 D6WNC9 A0A182K513 B4IYS0 A0A0Q9WVR9 A0A232FGG9 A0A1Q3FC69 A0A1Q3FCM6 K7J597 A0A0Q9XMU1 B0XEN9 A0A0Q9XRI0 A0A034WFN0 A0A034WD20 A0A0K8US19 A0A3B0JL47 B4MKJ9 A0A0V0G6S1 B5DPB6 A0A0A9WLA2 B4GRZ9 A0A1W4U6U0 B4QJK5 Q17MW1 Q9Y090 Q9VUC7 A0A182HCR9 B3ND11 B4HHC1 B3M3I2 B4PI00 A0A151WN25
A0A0L7QNM3 D6WNC9 A0A182K513 B4IYS0 A0A0Q9WVR9 A0A232FGG9 A0A1Q3FC69 A0A1Q3FCM6 K7J597 A0A0Q9XMU1 B0XEN9 A0A0Q9XRI0 A0A034WFN0 A0A034WD20 A0A0K8US19 A0A3B0JL47 B4MKJ9 A0A0V0G6S1 B5DPB6 A0A0A9WLA2 B4GRZ9 A0A1W4U6U0 B4QJK5 Q17MW1 Q9Y090 Q9VUC7 A0A182HCR9 B3ND11 B4HHC1 B3M3I2 B4PI00 A0A151WN25
PDB
5VCA
E-value=3.36225e-57,
Score=565
Ontologies
GO
PANTHER
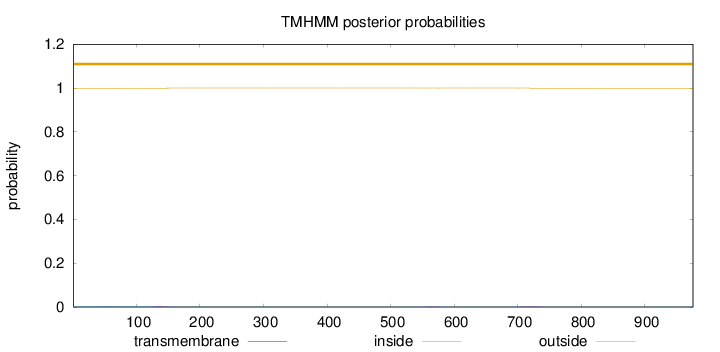
Topology
Length:
976
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0542
Exp number, first 60 AAs:
0.01235
Total prob of N-in:
0.00217
outside
1 - 976
Population Genetic Test Statistics
Pi
188.803965
Theta
152.655306
Tajima's D
0.927318
CLR
0.069922
CSRT
0.634818259087046
Interpretation
Possibly Positive selection
