Gene
KWMTBOMO02279
Pre Gene Modal
BGIBMGA003086
Annotation
PREDICTED:_leucine-rich_repeat-containing_G-protein_coupled_receptor_5_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.336 PlasmaMembrane Reliability : 1.914
Sequence
CDS
ATGGCGCCATTGCGATATGCCCACGTGATTTTTGCCTTCCTTGCCGCTAATTATATCATGGTGCTGTTGATGTTGCTACAACTACTCGCGTGGTGCAGTGGTGCGGCCGCTCTAGCGAACTGCCCAATAGGCTGCAACTGCAACGACGACACGCTCGTAGTGGTATGCGAGGAGAGCCGACTCGACGTACTACCGATAGCACTCAACCCATCCATCCAACGTTTAATTATCCGCAACAACAAAATCAAAACTATCGACAGCTCAATGCAGTTCTACACGGAACTCCAACACTTAGATCTCTCTCAGAATCACCTCGTGAATATACCAGTAAAAAGTTTTTCATATCAACGGAAATTGCAAGAATTGCATTTAAACCACAACAAAATATCGTCCGTGACGAACGCAACATTCCAAGGCTTGAACTCTTTGACCGTGCTGAATTTGAAACGCAACTTCTTACAGGAACTAACGAACGGCGTTTTCGCAACGTTGCCTAGACTTGAGGAGCTAAACTTGGGACAGAACAGCATCTCCAAAATCGATTCACGAGCCTTTGCGGGTCTTACGGCGCTTAGAATACTTTACTTGGATGATAACGCACTAAGTTCCGTGCCGACAGCATCATTCAGCCTCTTAGGCAGTCTAGCCGAACTCCACGTTGGTCTCAATGCGTTCTCATTCTTGCCCGATGATGCATTTGCTGGTTTGAACCGATTAACCGTACTAGACTTAAATGGAGCTGGGCTCTTTAACATTAGTGACAATGCGTTCAGAGGTCTTCCTGGACTTAGAAGTTTAAATCTCGTTGGCAACAGATTAAGTGTTGTACCGACACAACAATTAGGCGGTCTTACTAGATTGGAAGAACTATATATCGGTCAAAACGATTTTTTGACACTCGAAAGTCACTCTTTTAAGGGTCTAAAGAATTTGAAGTTAATCGATATAACAGGCGCAACCCAACTAGAAAGAGTAGAACGTGGAGCATTTGAAGACAACGTCAATTTGGAATCCGTGATTGTTGTAAATAATAAAAAGCTATCTGTGGTTGAAGAATCTGCACTCATTGGACTTTCTAAGCTCAAGCACGTTTCTTTGAGAGATAACGCAATAACAACACTAAGCGAATCTGTATTCGTTGGAAAAGAACTCAAACAGCTTGACATAACGGACAATCCTATATTCTGTGACTGTCAATTGTTGTGGCTACAAAGCCTTATGAATGACAAAAGCAACTTTACCCAAATCCAGTGCGCTAGTCCTGAACATTTAAAACACAAGTATTTGAGAACATTAAGCGCCGATGATCTGGGATGTGTACTCCACGATTCACGACAGCAGACTATCATTTGTATCGTGGTAGTAGCTTGCGTCGCCATATTAGCAACACTTGTTCTGATACTCTATAGATATCGAAAAAGTATGCAAGAAAAGCTAAAAGATTATAAATGGAATAAGGGACGTAAAAACTTAGACTACCACAAACCAATATCAACCGAAGAAGACTGCATCCTAGAGCTTTACGCCTCTCCGAGCGTGTTCTTCATGTCGGGCGGCCAGGGCTCCGCGCGCCGGCCGCACCGCTCGGGAGGCGGCGGTACGCTGCCCGCCAACGGCTTCACGTACATCGGTGGCGAGGGCCGGCCGCATCAACCACTCACCCTGAACAATGGCGCACCAGCCCATCATCTCAACAATGGCTCCTTGCGCTCCTTGCCCGATAAAAAGAACAACCGGAACGGCGTCGTTTGTCATCCAGAAAACTTCCAACGAAACCTGGACACTCGCTACTGTAGAAAACAAGAGAACGGGTACATACGCAATTCCGAAACGATAATCGGATTCCCGCGGAACGACCGGGAATACGAACGCGACCACGTGGAGTACAGTGAGCCAGAGTACTCTATCATCCCGGAGGCGTACGGCCGCGAGGAGTACCCGCGCTCCTGCTCGCACTCCAATACCTTCAACTGTTGA
Protein
MAPLRYAHVIFAFLAANYIMVLLMLLQLLAWCSGAAALANCPIGCNCNDDTLVVVCEESRLDVLPIALNPSIQRLIIRNNKIKTIDSSMQFYTELQHLDLSQNHLVNIPVKSFSYQRKLQELHLNHNKISSVTNATFQGLNSLTVLNLKRNFLQELTNGVFATLPRLEELNLGQNSISKIDSRAFAGLTALRILYLDDNALSSVPTASFSLLGSLAELHVGLNAFSFLPDDAFAGLNRLTVLDLNGAGLFNISDNAFRGLPGLRSLNLVGNRLSVVPTQQLGGLTRLEELYIGQNDFLTLESHSFKGLKNLKLIDITGATQLERVERGAFEDNVNLESVIVVNNKKLSVVEESALIGLSKLKHVSLRDNAITTLSESVFVGKELKQLDITDNPIFCDCQLLWLQSLMNDKSNFTQIQCASPEHLKHKYLRTLSADDLGCVLHDSRQQTIICIVVVACVAILATLVLILYRYRKSMQEKLKDYKWNKGRKNLDYHKPISTEEDCILELYASPSVFFMSGGQGSARRPHRSGGGGTLPANGFTYIGGEGRPHQPLTLNNGAPAHHLNNGSLRSLPDKKNNRNGVVCHPENFQRNLDTRYCRKQENGYIRNSETIIGFPRNDREYERDHVEYSEPEYSIIPEAYGREEYPRSCSHSNTFNC
Summary
Uniprot
H9J0P9
A0A212FGC2
A0A0L7LEF7
A0A2H1V633
A0A2A4JI45
A0A194PML1
+ More
D6WNE7 A0A0T6B8U7 A0A1Y1K6I9 N6TAN0 A0A2J7PFX4 A0A2P8YUM5 A0A067R1W3 A0A1W4X902 E0VEL8 T1H973 A0A182PPH6 A0A182N385 A0A182UFJ8 A0A1S4H6I9 A0A0A9XF14 A0A224XB29 A0A182QRV4 B0WLH0 A0A1B6E5V0 A0A1B6EGN5 A0A182HY11 J9K3S8 A0A2H8TEC7 A0A1B0DPB9 A0A182WXD5 A0A182LQ89 A0A182K4Z6 A0A1S4H7E0 A0A182J557 W5JQV0 A0A2M4CH83 A0A2M4BIR2 A0A2M4CG26 A0A182RR22 A0A2S2NZR6 A0A2M4BIP7 A0A2M3Z788 A0A182UXC7 A0A2M3Z7L6 A0A084WNV5 A0A2R7W5Q8 A0A182W2D6 A0A182YM23 A0A1B6MSC5 A0A182FBR1 E9IFU3 Q16K00 K7J4G7 A0A195DX90 A0A310SDJ4 A0A3L8DV46 A0A0J7L2C5 A0A195FPH3 A0A1B6JJ63 A0A195BCJ9 A0A158NJP2 F4X6E2 E2BH01 A0A088A4T2 A0A2A3EGZ6 A0A154P7P5 A0A0L7QVZ0 A0A151X691 A0A182G9J6 A0A336M8Y1 A0A0C9RFJ9 A0A0C9QTV1 A0A1B0AIB0 A0A1A9Y7C9 A0A1B0AU68 A0A1A9VQE9 A0A1B0G8L4 A0A0M4EI69 A0A0C9RTD4 A0A0A1WP62 A0A1A9WGQ7 B4LC16 A0A0M9ADC4 A0A0L0CFF7 A0A1A9WN41 A0A0K8V408 A0A034WJK2 A0A1I8MD81 A0A0L0BRR2 A0A1I8M752 Q9VU51 M9PFH7 A0A1I8P0K1 B4KZR4 A0A1A9YPR0
D6WNE7 A0A0T6B8U7 A0A1Y1K6I9 N6TAN0 A0A2J7PFX4 A0A2P8YUM5 A0A067R1W3 A0A1W4X902 E0VEL8 T1H973 A0A182PPH6 A0A182N385 A0A182UFJ8 A0A1S4H6I9 A0A0A9XF14 A0A224XB29 A0A182QRV4 B0WLH0 A0A1B6E5V0 A0A1B6EGN5 A0A182HY11 J9K3S8 A0A2H8TEC7 A0A1B0DPB9 A0A182WXD5 A0A182LQ89 A0A182K4Z6 A0A1S4H7E0 A0A182J557 W5JQV0 A0A2M4CH83 A0A2M4BIR2 A0A2M4CG26 A0A182RR22 A0A2S2NZR6 A0A2M4BIP7 A0A2M3Z788 A0A182UXC7 A0A2M3Z7L6 A0A084WNV5 A0A2R7W5Q8 A0A182W2D6 A0A182YM23 A0A1B6MSC5 A0A182FBR1 E9IFU3 Q16K00 K7J4G7 A0A195DX90 A0A310SDJ4 A0A3L8DV46 A0A0J7L2C5 A0A195FPH3 A0A1B6JJ63 A0A195BCJ9 A0A158NJP2 F4X6E2 E2BH01 A0A088A4T2 A0A2A3EGZ6 A0A154P7P5 A0A0L7QVZ0 A0A151X691 A0A182G9J6 A0A336M8Y1 A0A0C9RFJ9 A0A0C9QTV1 A0A1B0AIB0 A0A1A9Y7C9 A0A1B0AU68 A0A1A9VQE9 A0A1B0G8L4 A0A0M4EI69 A0A0C9RTD4 A0A0A1WP62 A0A1A9WGQ7 B4LC16 A0A0M9ADC4 A0A0L0CFF7 A0A1A9WN41 A0A0K8V408 A0A034WJK2 A0A1I8MD81 A0A0L0BRR2 A0A1I8M752 Q9VU51 M9PFH7 A0A1I8P0K1 B4KZR4 A0A1A9YPR0
Pubmed
19121390
22118469
26227816
26354079
18362917
19820115
+ More
28004739 23537049 29403074 24845553 20566863 12364791 25401762 20966253 20920257 23761445 24438588 25244985 21282665 17510324 20075255 30249741 21347285 21719571 20798317 26483478 25830018 17994087 26108605 25348373 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
28004739 23537049 29403074 24845553 20566863 12364791 25401762 20966253 20920257 23761445 24438588 25244985 21282665 17510324 20075255 30249741 21347285 21719571 20798317 26483478 25830018 17994087 26108605 25348373 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01010151
AGBW02008686
OWR52784.1
JTDY01001486
KOB73754.1
ODYU01000869
+ More
SOQ36277.1 NWSH01001395 PCG71388.1 KQ459598 KPI94557.1 KQ971343 EFA04362.1 LJIG01009096 KRT83763.1 GEZM01095864 JAV55295.1 APGK01044799 KB741028 KB632297 ENN74798.1 ERL91709.1 NEVH01025637 PNF15235.1 PYGN01000351 PSN47940.1 KK852762 KDR16993.1 DS235093 EEB11824.1 ACPB03001482 AAAB01008966 GBHO01027950 GBHO01027949 GBHO01027947 GBHO01026899 GBHO01024562 GBHO01024559 JAG15654.1 JAG15655.1 JAG15657.1 JAG16705.1 JAG19042.1 JAG19045.1 GFTR01006820 JAW09606.1 AXCN02001122 DS231985 EDS30475.1 GEDC01013545 GEDC01003987 GEDC01000630 JAS23753.1 JAS33311.1 JAS36668.1 GEDC01025799 GEDC01004068 GEDC01000248 JAS11499.1 JAS33230.1 JAS37050.1 APCN01004461 ABLF02019987 ABLF02019989 GFXV01000605 MBW12410.1 AJVK01018180 ADMH02000697 ETN65285.1 GGFL01000100 MBW64278.1 GGFJ01003796 MBW52937.1 GGFL01000095 MBW64273.1 GGMR01010005 MBY22624.1 GGFJ01003795 MBW52936.1 GGFM01003620 MBW24371.1 GGFM01003729 MBW24480.1 ATLV01024737 KE525359 KFB51899.1 KK854354 PTY15012.1 GEBQ01001136 JAT38841.1 GL762903 EFZ20530.1 CH477983 EAT34610.1 KQ980155 KYN17451.1 KQ762842 OAD55404.1 QOIP01000003 RLU24350.1 LBMM01001133 KMQ96851.1 KQ981424 KYN41844.1 GECU01024538 GECU01018251 GECU01013648 GECU01008554 JAS83168.1 JAS89455.1 JAS94058.1 JAS99152.1 KQ976526 KYM81947.1 ADTU01018143 ADTU01018144 ADTU01018145 ADTU01018146 ADTU01018147 ADTU01018148 ADTU01018149 ADTU01018150 ADTU01018151 ADTU01018152 GL888812 EGI57982.1 GL448255 EFN85034.1 KZ288251 PBC31045.1 KQ434835 KZC07892.1 KQ414716 KOC62798.1 KQ982482 KYQ55923.1 JXUM01049332 KQ561599 KXJ78041.1 UFQS01000566 UFQT01000566 SSX05007.1 SSX25369.1 GBYB01007165 JAG76932.1 GBYB01007166 GBYB01007169 JAG76933.1 JAG76936.1 JXJN01003538 CCAG010000270 CP012525 ALC44390.1 GBYB01012000 JAG81767.1 GBXI01013826 JAD00466.1 CH940647 EDW69816.2 KQ435696 KOX80829.1 JRES01000484 KNC30932.1 GDHF01029129 GDHF01018771 JAI23185.1 JAI33543.1 GAKP01004652 JAC54300.1 JRES01001456 KNC22716.1 AE014296 AY051439 AAF49839.1 AAK92863.1 AGB94489.1 CH933809 EDW19020.2
SOQ36277.1 NWSH01001395 PCG71388.1 KQ459598 KPI94557.1 KQ971343 EFA04362.1 LJIG01009096 KRT83763.1 GEZM01095864 JAV55295.1 APGK01044799 KB741028 KB632297 ENN74798.1 ERL91709.1 NEVH01025637 PNF15235.1 PYGN01000351 PSN47940.1 KK852762 KDR16993.1 DS235093 EEB11824.1 ACPB03001482 AAAB01008966 GBHO01027950 GBHO01027949 GBHO01027947 GBHO01026899 GBHO01024562 GBHO01024559 JAG15654.1 JAG15655.1 JAG15657.1 JAG16705.1 JAG19042.1 JAG19045.1 GFTR01006820 JAW09606.1 AXCN02001122 DS231985 EDS30475.1 GEDC01013545 GEDC01003987 GEDC01000630 JAS23753.1 JAS33311.1 JAS36668.1 GEDC01025799 GEDC01004068 GEDC01000248 JAS11499.1 JAS33230.1 JAS37050.1 APCN01004461 ABLF02019987 ABLF02019989 GFXV01000605 MBW12410.1 AJVK01018180 ADMH02000697 ETN65285.1 GGFL01000100 MBW64278.1 GGFJ01003796 MBW52937.1 GGFL01000095 MBW64273.1 GGMR01010005 MBY22624.1 GGFJ01003795 MBW52936.1 GGFM01003620 MBW24371.1 GGFM01003729 MBW24480.1 ATLV01024737 KE525359 KFB51899.1 KK854354 PTY15012.1 GEBQ01001136 JAT38841.1 GL762903 EFZ20530.1 CH477983 EAT34610.1 KQ980155 KYN17451.1 KQ762842 OAD55404.1 QOIP01000003 RLU24350.1 LBMM01001133 KMQ96851.1 KQ981424 KYN41844.1 GECU01024538 GECU01018251 GECU01013648 GECU01008554 JAS83168.1 JAS89455.1 JAS94058.1 JAS99152.1 KQ976526 KYM81947.1 ADTU01018143 ADTU01018144 ADTU01018145 ADTU01018146 ADTU01018147 ADTU01018148 ADTU01018149 ADTU01018150 ADTU01018151 ADTU01018152 GL888812 EGI57982.1 GL448255 EFN85034.1 KZ288251 PBC31045.1 KQ434835 KZC07892.1 KQ414716 KOC62798.1 KQ982482 KYQ55923.1 JXUM01049332 KQ561599 KXJ78041.1 UFQS01000566 UFQT01000566 SSX05007.1 SSX25369.1 GBYB01007165 JAG76932.1 GBYB01007166 GBYB01007169 JAG76933.1 JAG76936.1 JXJN01003538 CCAG010000270 CP012525 ALC44390.1 GBYB01012000 JAG81767.1 GBXI01013826 JAD00466.1 CH940647 EDW69816.2 KQ435696 KOX80829.1 JRES01000484 KNC30932.1 GDHF01029129 GDHF01018771 JAI23185.1 JAI33543.1 GAKP01004652 JAC54300.1 JRES01001456 KNC22716.1 AE014296 AY051439 AAF49839.1 AAK92863.1 AGB94489.1 CH933809 EDW19020.2
Proteomes
UP000005204
UP000007151
UP000037510
UP000218220
UP000053268
UP000007266
+ More
UP000019118 UP000030742 UP000235965 UP000245037 UP000027135 UP000192223 UP000009046 UP000015103 UP000075885 UP000075884 UP000075902 UP000075886 UP000002320 UP000075840 UP000007819 UP000092462 UP000076407 UP000075882 UP000075881 UP000075880 UP000000673 UP000075900 UP000075903 UP000030765 UP000075920 UP000076408 UP000069272 UP000008820 UP000002358 UP000078492 UP000279307 UP000036403 UP000078541 UP000078540 UP000005205 UP000007755 UP000008237 UP000005203 UP000242457 UP000076502 UP000053825 UP000075809 UP000069940 UP000249989 UP000092445 UP000092443 UP000092460 UP000078200 UP000092444 UP000092553 UP000091820 UP000008792 UP000053105 UP000037069 UP000095301 UP000000803 UP000095300 UP000009192
UP000019118 UP000030742 UP000235965 UP000245037 UP000027135 UP000192223 UP000009046 UP000015103 UP000075885 UP000075884 UP000075902 UP000075886 UP000002320 UP000075840 UP000007819 UP000092462 UP000076407 UP000075882 UP000075881 UP000075880 UP000000673 UP000075900 UP000075903 UP000030765 UP000075920 UP000076408 UP000069272 UP000008820 UP000002358 UP000078492 UP000279307 UP000036403 UP000078541 UP000078540 UP000005205 UP000007755 UP000008237 UP000005203 UP000242457 UP000076502 UP000053825 UP000075809 UP000069940 UP000249989 UP000092445 UP000092443 UP000092460 UP000078200 UP000092444 UP000092553 UP000091820 UP000008792 UP000053105 UP000037069 UP000095301 UP000000803 UP000095300 UP000009192
Interpro
Gene 3D
ProteinModelPortal
H9J0P9
A0A212FGC2
A0A0L7LEF7
A0A2H1V633
A0A2A4JI45
A0A194PML1
+ More
D6WNE7 A0A0T6B8U7 A0A1Y1K6I9 N6TAN0 A0A2J7PFX4 A0A2P8YUM5 A0A067R1W3 A0A1W4X902 E0VEL8 T1H973 A0A182PPH6 A0A182N385 A0A182UFJ8 A0A1S4H6I9 A0A0A9XF14 A0A224XB29 A0A182QRV4 B0WLH0 A0A1B6E5V0 A0A1B6EGN5 A0A182HY11 J9K3S8 A0A2H8TEC7 A0A1B0DPB9 A0A182WXD5 A0A182LQ89 A0A182K4Z6 A0A1S4H7E0 A0A182J557 W5JQV0 A0A2M4CH83 A0A2M4BIR2 A0A2M4CG26 A0A182RR22 A0A2S2NZR6 A0A2M4BIP7 A0A2M3Z788 A0A182UXC7 A0A2M3Z7L6 A0A084WNV5 A0A2R7W5Q8 A0A182W2D6 A0A182YM23 A0A1B6MSC5 A0A182FBR1 E9IFU3 Q16K00 K7J4G7 A0A195DX90 A0A310SDJ4 A0A3L8DV46 A0A0J7L2C5 A0A195FPH3 A0A1B6JJ63 A0A195BCJ9 A0A158NJP2 F4X6E2 E2BH01 A0A088A4T2 A0A2A3EGZ6 A0A154P7P5 A0A0L7QVZ0 A0A151X691 A0A182G9J6 A0A336M8Y1 A0A0C9RFJ9 A0A0C9QTV1 A0A1B0AIB0 A0A1A9Y7C9 A0A1B0AU68 A0A1A9VQE9 A0A1B0G8L4 A0A0M4EI69 A0A0C9RTD4 A0A0A1WP62 A0A1A9WGQ7 B4LC16 A0A0M9ADC4 A0A0L0CFF7 A0A1A9WN41 A0A0K8V408 A0A034WJK2 A0A1I8MD81 A0A0L0BRR2 A0A1I8M752 Q9VU51 M9PFH7 A0A1I8P0K1 B4KZR4 A0A1A9YPR0
D6WNE7 A0A0T6B8U7 A0A1Y1K6I9 N6TAN0 A0A2J7PFX4 A0A2P8YUM5 A0A067R1W3 A0A1W4X902 E0VEL8 T1H973 A0A182PPH6 A0A182N385 A0A182UFJ8 A0A1S4H6I9 A0A0A9XF14 A0A224XB29 A0A182QRV4 B0WLH0 A0A1B6E5V0 A0A1B6EGN5 A0A182HY11 J9K3S8 A0A2H8TEC7 A0A1B0DPB9 A0A182WXD5 A0A182LQ89 A0A182K4Z6 A0A1S4H7E0 A0A182J557 W5JQV0 A0A2M4CH83 A0A2M4BIR2 A0A2M4CG26 A0A182RR22 A0A2S2NZR6 A0A2M4BIP7 A0A2M3Z788 A0A182UXC7 A0A2M3Z7L6 A0A084WNV5 A0A2R7W5Q8 A0A182W2D6 A0A182YM23 A0A1B6MSC5 A0A182FBR1 E9IFU3 Q16K00 K7J4G7 A0A195DX90 A0A310SDJ4 A0A3L8DV46 A0A0J7L2C5 A0A195FPH3 A0A1B6JJ63 A0A195BCJ9 A0A158NJP2 F4X6E2 E2BH01 A0A088A4T2 A0A2A3EGZ6 A0A154P7P5 A0A0L7QVZ0 A0A151X691 A0A182G9J6 A0A336M8Y1 A0A0C9RFJ9 A0A0C9QTV1 A0A1B0AIB0 A0A1A9Y7C9 A0A1B0AU68 A0A1A9VQE9 A0A1B0G8L4 A0A0M4EI69 A0A0C9RTD4 A0A0A1WP62 A0A1A9WGQ7 B4LC16 A0A0M9ADC4 A0A0L0CFF7 A0A1A9WN41 A0A0K8V408 A0A034WJK2 A0A1I8MD81 A0A0L0BRR2 A0A1I8M752 Q9VU51 M9PFH7 A0A1I8P0K1 B4KZR4 A0A1A9YPR0
PDB
4U7L
E-value=1.29561e-32,
Score=351
Ontologies
KEGG
GO
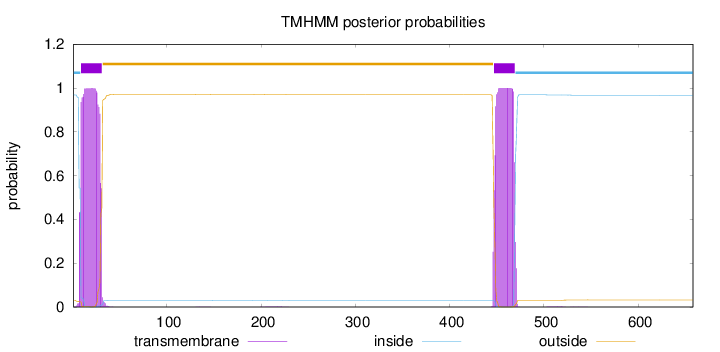
Topology
Length:
658
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.1784099999999
Exp number, first 60 AAs:
22.63374
Total prob of N-in:
0.96959
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 31
outside
32 - 446
TMhelix
447 - 469
inside
470 - 658
Population Genetic Test Statistics
Pi
27.093467
Theta
189.152091
Tajima's D
0.745157
CLR
0.728389
CSRT
0.588270586470676
Interpretation
Uncertain
