Gene
KWMTBOMO02257 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003057
Annotation
alpha_amylase_precursor_[Bombyx_mori]
Full name
Maltase A1
+ More
Maltase 2
Maltase 2
Alternative Name
Larval visceral protein H
Location in the cell
Cytoplasmic Reliability : 3.381
Sequence
CDS
ATGAGTGCGAAATGGAGAATTGGAATTATTTTACTGCCGGTGCTGGTTTTAGTACAGTGTCAAGTAAAAGAATTAGATTGGTGGGAAAAGACCGTTTTCTATCAAATTTACCCTAGGTCTTTCAAAGACAGTGACGGTGATGGTATTGGAGACCTGAATGGAATAACAGATAGCTTAGAATATCTAAAGGAATTGGGTGTTGGTGCGACATGGCTATCACCGATTTTCAAATCGCCAATGTACGATTTTGGATACGACATTTCTGATTTTTATGCTATCCAAGAAGAATATGGTACAATGGAAGACTTTGAGAGGCTTTTACAAAAAGCAAAAGAGTTGGATATTAAAATTGTTCTTGATTTCGTACCTAACCACGGCAGTAACGAAAGTGTTTGGTTTGAAGAAGCTCTCAAAGGACACGAAAAATATTACAATTACTTTGTCTGGGAAGACGGAGTTGAAGATGAAAACGGAAAATTAAATCCTCCCAACAACTGGGTCAGCGTATTCCGAAAAAGTGCTTGGGAATACAGAAAAGAGGTGGGTAAATACTATCTTCATCAGTTTGTTATCGGACAGCCTGATTTAAATTACCGCAATCCAGAGGTGGTTGAAGAAATGAAGAACGTTATTCGCTTCTGGCTCGATAAGGGTATTGCTGGATTCAGGGTGGATGCGATCGCTCATTTATTTGAAGTAGACAAAACACTGTTTGGAGGTAAATATCCTGATGAGCCATTGGCACAAAATAGAGAGATCGATCCAGACAATTATGACTATTTAGATCACATCTACACAAAAGACCACGAAGAAACATTTGAAATGGTTTACCAATGGAGAGACGTTCTTGATGAGTATAAAGCAAAAGATGGATTCACTAGGGTTATGATGACTGAAGCTTACTCCAGTCCTCAGATCACAATGAAGTATTTTGGTGATGGTGTCCGTGCTGGAGCTCAAATGCCATTCAATTTCGTACTCATTTCCGAAGTTAGTGGTTCATCTACTGCTGCAGAGCTTAAATACGCTTTGGACAAATTTTTGACCTTTAAGCCTGTGGACAAGTTGGCTAACTGGGTGGCTGGTAATCACGACAACAACCGAGTAGCTTCTAGGTATAGTGTTGAGCTAGTAGATGGGCTTAATATGATTGTTATGCTTCTACCTGGTATAGCTGTAACTTATATGGGCGAAGAAATCGGAATGGTTGACGGATACGTAAGCTGGGAAGATACGGTTGACCCCAGTGGATGTAACACAGATGATCCTATCAACTACTGGACTGTTTCTAGAGACCCTGAACGCACACCATTTCAATGGAACTCTGAGAAAAATGCTGGATTCTCTACTGGTGATAAAACATGGCTGCCAGTAGCGGAGGGCTATGAAACCCTCAATGTCGAAATTCAAAAATCGACTTCTCGCTCTCACTTGAACGTTTACAAGCAACTGACACGTCTGCGAAATGAACCTGTTTTCCGGTATGGTCGCTTCGAATCAGTGGCACTTAACCCTGATATTTTCGCTTTCAAAAGATGGCACGATGGTGAAATTTACGTGACAGTGATCAATTTGAAGAATCGTGATCATACTATTGACTTAACTTACTTCGAGCATGTTGTTGGTCATTTAGAAGTTGTCCTTACTGATATTCGTTCCAAGAAGGTTCCTGGAGATTTGCTGGAAGCGAGCCAAGCGGAGATCGTCGCCCACGAATCTTTAGTGCTCAAAGTTGTACACACCCTACGGTAA
Protein
MSAKWRIGIILLPVLVLVQCQVKELDWWEKTVFYQIYPRSFKDSDGDGIGDLNGITDSLEYLKELGVGATWLSPIFKSPMYDFGYDISDFYAIQEEYGTMEDFERLLQKAKELDIKIVLDFVPNHGSNESVWFEEALKGHEKYYNYFVWEDGVEDENGKLNPPNNWVSVFRKSAWEYRKEVGKYYLHQFVIGQPDLNYRNPEVVEEMKNVIRFWLDKGIAGFRVDAIAHLFEVDKTLFGGKYPDEPLAQNREIDPDNYDYLDHIYTKDHEETFEMVYQWRDVLDEYKAKDGFTRVMMTEAYSSPQITMKYFGDGVRAGAQMPFNFVLISEVSGSSTAAELKYALDKFLTFKPVDKLANWVAGNHDNNRVASRYSVELVDGLNMIVMLLPGIAVTYMGEEIGMVDGYVSWEDTVDPSGCNTDDPINYWTVSRDPERTPFQWNSEKNAGFSTGDKTWLPVAEGYETLNVEIQKSTSRSHLNVYKQLTRLRNEPVFRYGRFESVALNPDIFAFKRWHDGEIYVTVINLKNRDHTIDLTYFEHVVGHLEVVLTDIRSKKVPGDLLEASQAEIVAHESLVLKVVHTLR
Summary
Catalytic Activity
Hydrolysis of terminal, non-reducing (1->4)-linked alpha-D-glucose residues with release of alpha-D-glucose.
Similarity
Belongs to the glycosyl hydrolase 13 family.
Keywords
Complete proteome
Glycoprotein
Glycosidase
Hydrolase
Reference proteome
Signal
Feature
chain Maltase A1
Uniprot
D8KY55
A0A0D3R469
A0A0N0PB16
A0A194PPD0
A0A2H1VQK1
A0A194PTX8
+ More
A0A2W1BJR6 A0A2A4JP22 A0A212F5X2 A0A2W1BM09 A0A2A4JQ74 A0A0N0PAR2 H9J0L9 A0A212F5V9 A0A0L7LPH9 A0A2W1BRC7 A0A2A4JNL4 H9J0L8 A0A0N1INK8 A0A2H1V8G1 A0A212F5W6 B4J5W7 A0A2A4JCN0 A0A2H1W014 A0A212F6K0 B4KP60 B4LLB2 B4P2T8 B5DZA0 T1PAD3 A0A1I8MF97 B3N8P3 B4HRX5 B4QFR9 B3MFY2 A0A1W4UYX9 B4MQ26 P07190 A0A0J9TWA3 A0A0A1X1H1 A0A2A4JCK8 A0A3B0JKC9 A0A034WCA5 A0A0L0CQC7 A0A0M5IX79 A0A1I8M6J4 B4LLB7 A0A1B0DP22 B7T4S1 W5JEB7 A0A1I8Q4H4 W8B7U7 A0A0N1I9T4 A0A1L8EB24 B4KIU7 O16099 A0A194QDR1 A0A1L8EBH7 Q292I4 B4MQ22 A0A1L8EBG3 B4KP53 A0A0M3QUM5 A0A0A1XB46 A0A1L8EBF6 U5EJL5 W8ATZ7 B4GDF4 A0A1L8EBM5 A0A1L8EBB5 W8AQ09 B4GDE7 A0A182G0E9 A0A2C9GRJ1 Q29LL3 A0A182JIX4 B4MQ20 A0A3G5BIE7 A0A2M4BI09 A0A182LQZ3 B5DZA1 A0A0R3NTH9 B4KP55 A0A182UTX9 U5EX75 Q7PYT9 B4LLB9 A0A1I8N8F6 B4GDF0 B4J5X2 A0A3B0JM24 B5DZA2 A0A182LUR9 B4KP54 A0A0Q9XGB8 A0A1S4FQI8 A0A336M5S6 A0A1L8DR17 A0A0Q9WLB7 A0A3B0JRB9
A0A2W1BJR6 A0A2A4JP22 A0A212F5X2 A0A2W1BM09 A0A2A4JQ74 A0A0N0PAR2 H9J0L9 A0A212F5V9 A0A0L7LPH9 A0A2W1BRC7 A0A2A4JNL4 H9J0L8 A0A0N1INK8 A0A2H1V8G1 A0A212F5W6 B4J5W7 A0A2A4JCN0 A0A2H1W014 A0A212F6K0 B4KP60 B4LLB2 B4P2T8 B5DZA0 T1PAD3 A0A1I8MF97 B3N8P3 B4HRX5 B4QFR9 B3MFY2 A0A1W4UYX9 B4MQ26 P07190 A0A0J9TWA3 A0A0A1X1H1 A0A2A4JCK8 A0A3B0JKC9 A0A034WCA5 A0A0L0CQC7 A0A0M5IX79 A0A1I8M6J4 B4LLB7 A0A1B0DP22 B7T4S1 W5JEB7 A0A1I8Q4H4 W8B7U7 A0A0N1I9T4 A0A1L8EB24 B4KIU7 O16099 A0A194QDR1 A0A1L8EBH7 Q292I4 B4MQ22 A0A1L8EBG3 B4KP53 A0A0M3QUM5 A0A0A1XB46 A0A1L8EBF6 U5EJL5 W8ATZ7 B4GDF4 A0A1L8EBM5 A0A1L8EBB5 W8AQ09 B4GDE7 A0A182G0E9 A0A2C9GRJ1 Q29LL3 A0A182JIX4 B4MQ20 A0A3G5BIE7 A0A2M4BI09 A0A182LQZ3 B5DZA1 A0A0R3NTH9 B4KP55 A0A182UTX9 U5EX75 Q7PYT9 B4LLB9 A0A1I8N8F6 B4GDF0 B4J5X2 A0A3B0JM24 B5DZA2 A0A182LUR9 B4KP54 A0A0Q9XGB8 A0A1S4FQI8 A0A336M5S6 A0A1L8DR17 A0A0Q9WLB7 A0A3B0JRB9
EC Number
3.2.1.20
Pubmed
EMBL
BABH01010222
EU856718
ACJ49024.1
KP101252
AJR29308.1
KQ461183
+ More
KPJ07577.1 KQ459598 KPI94589.1 ODYU01003827 SOQ43077.1 KPI94590.1 KZ150000 PZC75322.1 NWSH01000913 PCG73559.1 AGBW02010122 OWR49124.1 PZC75321.1 PCG73560.1 KPJ07576.1 OWR49125.1 JTDY01000407 KOB77337.1 PZC75320.1 PCG73561.1 BABH01010223 KPJ07575.1 ODYU01001035 SOQ36692.1 OWR49126.1 CH916367 EDW00810.1 NWSH01001872 PCG69835.1 ODYU01005517 SOQ46439.1 AGBW02010006 OWR49357.1 CH933808 EDW10126.1 CH940648 EDW60849.1 CM000157 EDW89349.2 CM000071 EDY68786.1 KA645654 AFP60283.1 CH954177 EDV59520.2 CH480816 EDW46938.1 CM000362 EDX06175.1 CH902619 EDV35664.1 CH963849 EDW74215.1 V00204 AE013599 AY070626 CM002911 KMY92265.1 GBXI01009571 JAD04721.1 PCG69837.1 OUUW01000001 SPP73696.1 GAKP01007519 JAC51433.1 JRES01000171 KNC33639.1 CP012524 ALC40924.1 EDW60854.1 AJVK01007891 AJVK01007892 EU934324 ACI30102.1 ADMH02001754 ETN61169.1 GAMC01013442 JAB93113.1 KQ460041 KPJ18359.1 GFDG01002842 JAV15957.1 CH933807 EDW12453.2 CH940649 AF006573 KQ459324 KPJ01591.1 GFDG01002843 JAV15956.1 EAL24878.2 EDW74211.1 GFDG01002844 JAV15955.1 EDW10119.1 ALC40929.1 GBXI01005733 JAD08559.1 GFDG01002847 JAV15952.1 GANO01002192 JAB57679.1 GAMC01018417 JAB88138.1 CH479181 EDW31630.1 GFDG01002846 JAV15953.1 GFDG01002845 JAV15954.1 GAMC01018418 JAB88137.1 EDW31623.1 APCN01002016 CH379060 EAL34032.3 EDW74209.1 MK075157 AYV99560.1 GGFJ01003536 MBW52677.1 EDY68787.1 KRT04418.1 EDW10121.1 GANO01002670 JAB57201.1 AAAB01008987 EAA00998.3 EDW60856.1 EDW31626.1 EDW00815.1 SPP73691.1 EDY68788.2 AXCM01004815 EDW10120.1 KRG03249.1 UFQT01000190 SSX21388.1 GFDF01005270 JAV08814.1 KRF81627.1 SPP73688.1
KPJ07577.1 KQ459598 KPI94589.1 ODYU01003827 SOQ43077.1 KPI94590.1 KZ150000 PZC75322.1 NWSH01000913 PCG73559.1 AGBW02010122 OWR49124.1 PZC75321.1 PCG73560.1 KPJ07576.1 OWR49125.1 JTDY01000407 KOB77337.1 PZC75320.1 PCG73561.1 BABH01010223 KPJ07575.1 ODYU01001035 SOQ36692.1 OWR49126.1 CH916367 EDW00810.1 NWSH01001872 PCG69835.1 ODYU01005517 SOQ46439.1 AGBW02010006 OWR49357.1 CH933808 EDW10126.1 CH940648 EDW60849.1 CM000157 EDW89349.2 CM000071 EDY68786.1 KA645654 AFP60283.1 CH954177 EDV59520.2 CH480816 EDW46938.1 CM000362 EDX06175.1 CH902619 EDV35664.1 CH963849 EDW74215.1 V00204 AE013599 AY070626 CM002911 KMY92265.1 GBXI01009571 JAD04721.1 PCG69837.1 OUUW01000001 SPP73696.1 GAKP01007519 JAC51433.1 JRES01000171 KNC33639.1 CP012524 ALC40924.1 EDW60854.1 AJVK01007891 AJVK01007892 EU934324 ACI30102.1 ADMH02001754 ETN61169.1 GAMC01013442 JAB93113.1 KQ460041 KPJ18359.1 GFDG01002842 JAV15957.1 CH933807 EDW12453.2 CH940649 AF006573 KQ459324 KPJ01591.1 GFDG01002843 JAV15956.1 EAL24878.2 EDW74211.1 GFDG01002844 JAV15955.1 EDW10119.1 ALC40929.1 GBXI01005733 JAD08559.1 GFDG01002847 JAV15952.1 GANO01002192 JAB57679.1 GAMC01018417 JAB88138.1 CH479181 EDW31630.1 GFDG01002846 JAV15953.1 GFDG01002845 JAV15954.1 GAMC01018418 JAB88137.1 EDW31623.1 APCN01002016 CH379060 EAL34032.3 EDW74209.1 MK075157 AYV99560.1 GGFJ01003536 MBW52677.1 EDY68787.1 KRT04418.1 EDW10121.1 GANO01002670 JAB57201.1 AAAB01008987 EAA00998.3 EDW60856.1 EDW31626.1 EDW00815.1 SPP73691.1 EDY68788.2 AXCM01004815 EDW10120.1 KRG03249.1 UFQT01000190 SSX21388.1 GFDF01005270 JAV08814.1 KRF81627.1 SPP73688.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000037510
+ More
UP000001070 UP000009192 UP000008792 UP000002282 UP000001819 UP000095301 UP000008711 UP000001292 UP000000304 UP000007801 UP000192221 UP000007798 UP000000803 UP000268350 UP000037069 UP000092553 UP000092462 UP000000673 UP000095300 UP000008744 UP000069272 UP000075840 UP000075880 UP000075882 UP000075903 UP000007062 UP000075883
UP000001070 UP000009192 UP000008792 UP000002282 UP000001819 UP000095301 UP000008711 UP000001292 UP000000304 UP000007801 UP000192221 UP000007798 UP000000803 UP000268350 UP000037069 UP000092553 UP000092462 UP000000673 UP000095300 UP000008744 UP000069272 UP000075840 UP000075880 UP000075882 UP000075903 UP000007062 UP000075883
Pfam
PF00128 Alpha-amylase
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
D8KY55
A0A0D3R469
A0A0N0PB16
A0A194PPD0
A0A2H1VQK1
A0A194PTX8
+ More
A0A2W1BJR6 A0A2A4JP22 A0A212F5X2 A0A2W1BM09 A0A2A4JQ74 A0A0N0PAR2 H9J0L9 A0A212F5V9 A0A0L7LPH9 A0A2W1BRC7 A0A2A4JNL4 H9J0L8 A0A0N1INK8 A0A2H1V8G1 A0A212F5W6 B4J5W7 A0A2A4JCN0 A0A2H1W014 A0A212F6K0 B4KP60 B4LLB2 B4P2T8 B5DZA0 T1PAD3 A0A1I8MF97 B3N8P3 B4HRX5 B4QFR9 B3MFY2 A0A1W4UYX9 B4MQ26 P07190 A0A0J9TWA3 A0A0A1X1H1 A0A2A4JCK8 A0A3B0JKC9 A0A034WCA5 A0A0L0CQC7 A0A0M5IX79 A0A1I8M6J4 B4LLB7 A0A1B0DP22 B7T4S1 W5JEB7 A0A1I8Q4H4 W8B7U7 A0A0N1I9T4 A0A1L8EB24 B4KIU7 O16099 A0A194QDR1 A0A1L8EBH7 Q292I4 B4MQ22 A0A1L8EBG3 B4KP53 A0A0M3QUM5 A0A0A1XB46 A0A1L8EBF6 U5EJL5 W8ATZ7 B4GDF4 A0A1L8EBM5 A0A1L8EBB5 W8AQ09 B4GDE7 A0A182G0E9 A0A2C9GRJ1 Q29LL3 A0A182JIX4 B4MQ20 A0A3G5BIE7 A0A2M4BI09 A0A182LQZ3 B5DZA1 A0A0R3NTH9 B4KP55 A0A182UTX9 U5EX75 Q7PYT9 B4LLB9 A0A1I8N8F6 B4GDF0 B4J5X2 A0A3B0JM24 B5DZA2 A0A182LUR9 B4KP54 A0A0Q9XGB8 A0A1S4FQI8 A0A336M5S6 A0A1L8DR17 A0A0Q9WLB7 A0A3B0JRB9
A0A2W1BJR6 A0A2A4JP22 A0A212F5X2 A0A2W1BM09 A0A2A4JQ74 A0A0N0PAR2 H9J0L9 A0A212F5V9 A0A0L7LPH9 A0A2W1BRC7 A0A2A4JNL4 H9J0L8 A0A0N1INK8 A0A2H1V8G1 A0A212F5W6 B4J5W7 A0A2A4JCN0 A0A2H1W014 A0A212F6K0 B4KP60 B4LLB2 B4P2T8 B5DZA0 T1PAD3 A0A1I8MF97 B3N8P3 B4HRX5 B4QFR9 B3MFY2 A0A1W4UYX9 B4MQ26 P07190 A0A0J9TWA3 A0A0A1X1H1 A0A2A4JCK8 A0A3B0JKC9 A0A034WCA5 A0A0L0CQC7 A0A0M5IX79 A0A1I8M6J4 B4LLB7 A0A1B0DP22 B7T4S1 W5JEB7 A0A1I8Q4H4 W8B7U7 A0A0N1I9T4 A0A1L8EB24 B4KIU7 O16099 A0A194QDR1 A0A1L8EBH7 Q292I4 B4MQ22 A0A1L8EBG3 B4KP53 A0A0M3QUM5 A0A0A1XB46 A0A1L8EBF6 U5EJL5 W8ATZ7 B4GDF4 A0A1L8EBM5 A0A1L8EBB5 W8AQ09 B4GDE7 A0A182G0E9 A0A2C9GRJ1 Q29LL3 A0A182JIX4 B4MQ20 A0A3G5BIE7 A0A2M4BI09 A0A182LQZ3 B5DZA1 A0A0R3NTH9 B4KP55 A0A182UTX9 U5EX75 Q7PYT9 B4LLB9 A0A1I8N8F6 B4GDF0 B4J5X2 A0A3B0JM24 B5DZA2 A0A182LUR9 B4KP54 A0A0Q9XGB8 A0A1S4FQI8 A0A336M5S6 A0A1L8DR17 A0A0Q9WLB7 A0A3B0JRB9
PDB
5ZCE
E-value=2.35837e-76,
Score=728
Ontologies
GO
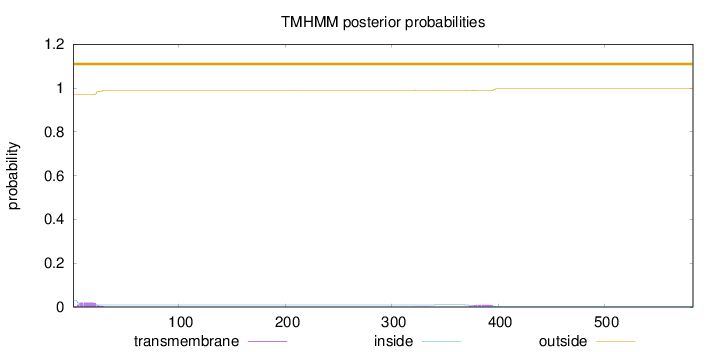
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.979352

Length:
583
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.59132
Exp number, first 60 AAs:
0.37543
Total prob of N-in:
0.02904
outside
1 - 583
Population Genetic Test Statistics
Pi
265.128829
Theta
203.251787
Tajima's D
0.950217
CLR
0
CSRT
0.650417479126044
Interpretation
Uncertain
