Gene
KWMTBOMO02256 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003056
Annotation
PREDICTED:_maltase_A1-like_[Amyelois_transitella]
Full name
Maltase 2
+ More
Maltase A1
Maltase A1
Alternative Name
Larval visceral protein H
Location in the cell
Cytoplasmic Reliability : 1.495
Sequence
CDS
ATGAAGACAGTGTGTTTGCTGTCTTTGCTTTTCGTTGCATGTAGTGGAATCATTATAAAGAATGGAGAAGTTCAAGATTGGTGGGAGACTTCAATTTTATACCAGATCTACCCAAGGTCTTTTGCTGATAGCGACGGCGATGGTATTGGCGATTTGAATGGTATTACTTCAAAATTGGAATATATCAAAGAATTAGGTGTAGGAGCTGTTTGGCTTTCGCCGATTTTCAAATCGCCCATGGTTGATTTTGGCTACGACATTGCTAATTTTTATGAAATCCACCATGAATATGGTACGATGGAAGATTTCGAAGCTCTATTGAAAAAGGCGAATGAATTAGACATCAAAGTTGTTTTGGATCTTGTACCAAATCATACATCAAACGAAAGTGTTTGGTTCCAAGAAGCTCTCAATGGCAACGAAAAATATTACAATTATTTCGTTTGGGAAGACGGAATCATAGACGAAAATGGAAATAGGCAGCCACCGAATAATTGGTTGAGTCACTTTAGAGGCAGCGCTTGGGAATACAAAGAAGAAGTCGGAAAATATTACTTACATCAATTCGCTGTTGGACAGCCAGACTTGAACTACCGCAATCAAGATGTCGTAGATGAAATGAAAAACATAATCCGGTTCTGGCTTGGAAAAGGTATTGCCGGATTCAGGGTAGATGCCGTGAATTGTCTCTACGAAGCCGACAAGAATCTGTATGGTGGAAAGTACCCTGATGAACCCTTATCTGGTAGACTGGATGTTGACTCAGAATCCCATGATTACCTGCAACATATTCACACCAAGGACCACAACGACACTTACTATATGGTTTACCAATGGAGAGATGTCTTCGATGAGTTCAAGGCGATCGATGGGCTGACTAGAGCTATGATGACCGAAGTATATGCATCGATCCAAGATGTGGTTAGGTATTTCGGTGAAGGAGATTTAAAAGGAGCACAGATTCCGTTTAATTTCGATTTGATCACTGACGTTGATGCCAGTTCATCAGCTGCTGATATAAAAAGAGCCGTTGATAAGTTCTTAACTTATAAACCAGTCGACCAAACAGCTAATTGGGTGGTTGGGAATCACGACAATAGCCGAATGGCTACAAGATATGGTGCTTCATTGGTAGATGGTATCAACATGCTAGTATTACTTCTTCCGGGAGTAGCAGTTACGTATATGGGAGAAGAAATTGGACTGGAAGACGGCTATGTTAGCTGGGAAGACACAGTTGACCCATCTGGATGTAATACCAACGACCCAATAAAATATGTCGAATCGTCGAGAGATCCCGAAAGGACTCCTTTCCATTGGAACCCCGAGAAAAACGCTGGATTCTCTACCGCTGACAAGACCTGGTTGCCGATGGCCGAGGGCTATGAAACTTTGAACGTTGAAGTTCAGAAGGCTTCAGAAAGGTCCCATCTTAAAGTTTACAAGGCTTTGTCAGATTTACGTCAAGAAAACACATTCAGATACGGCCGTTATGAGTCTTTAGCTTTAAATCAGGATATTTTTGTATTTAAAAGATGGCTTAACGACGTCATTTATCTAGTGGTTGTGAATATGCGAGACGTGGAACATAACATTGATCTTACATACTTCGAAAATGTTTCTGGGAACGTTGCCGTGTCGATAAGGAGCGTTAACTCACCTAAAAATGAGGGGGATACATTCGACGCGAAATCTCTACCCGTTGTCGGATTCGAAGGACTCGTCTTGAAGGTTGTTTAG
Protein
MKTVCLLSLLFVACSGIIIKNGEVQDWWETSILYQIYPRSFADSDGDGIGDLNGITSKLEYIKELGVGAVWLSPIFKSPMVDFGYDIANFYEIHHEYGTMEDFEALLKKANELDIKVVLDLVPNHTSNESVWFQEALNGNEKYYNYFVWEDGIIDENGNRQPPNNWLSHFRGSAWEYKEEVGKYYLHQFAVGQPDLNYRNQDVVDEMKNIIRFWLGKGIAGFRVDAVNCLYEADKNLYGGKYPDEPLSGRLDVDSESHDYLQHIHTKDHNDTYYMVYQWRDVFDEFKAIDGLTRAMMTEVYASIQDVVRYFGEGDLKGAQIPFNFDLITDVDASSSAADIKRAVDKFLTYKPVDQTANWVVGNHDNSRMATRYGASLVDGINMLVLLLPGVAVTYMGEEIGLEDGYVSWEDTVDPSGCNTNDPIKYVESSRDPERTPFHWNPEKNAGFSTADKTWLPMAEGYETLNVEVQKASERSHLKVYKALSDLRQENTFRYGRYESLALNQDIFVFKRWLNDVIYLVVVNMRDVEHNIDLTYFENVSGNVAVSIRSVNSPKNEGDTFDAKSLPVVGFEGLVLKVV
Summary
Catalytic Activity
Hydrolysis of terminal, non-reducing (1->4)-linked alpha-D-glucose residues with release of alpha-D-glucose.
Similarity
Belongs to the glycosyl hydrolase 13 family.
Keywords
Complete proteome
Glycoprotein
Glycosidase
Hydrolase
Reference proteome
Signal
Feature
chain Maltase 2
Uniprot
H9J0L9
A0A2A4JQ74
A0A212F5V9
A0A2W1BM09
A0A2H1VQK1
A0A0N0PAR2
+ More
A0A194PTX8 A0A194PPD0 A0A0D3R469 A0A0N0PB16 D8KY55 A0A2W1BJR6 A0A2A4JP22 A0A212F5X2 A0A0L7LPH9 A0A2W1BRC7 A0A2A4JNL4 H9J0L8 A0A2H1V8G1 A0A0N1INK8 A0A212F5W6 B4LLB9 B4KP53 Q292I4 W8ATZ7 W8AQ09 B4GDF4 A0A0N1I9T4 B4MQ20 A0A0A1XB46 A0A034WCA5 B4J5W7 A0A194QDR1 B3N8N6 A0A1I8N8F6 A0A3B0JRB9 B3MFX5 B4QFT0 A0A0J9R808 H5V882 A1Z7F3 A0A0M5J9Z6 A0A212F6K0 E0R939 B4J5X4 A0A1I8MGR6 B4KP60 T1PFH0 A0A3B0JJS3 A0A0L0CNA2 A0A1I8M319 B4P2U5 A0A1W4UXI6 A0A1W4UKB0 A0A212F7W3 B4MQ26 B4GDF3 B3MFY2 B3N8N7 A0A2H1W014 U5ELE7 A0A0M5IX79 A0A1I8PZX7 A0A0J9R8A6 A1Z7F2 A0A2A4JCN0 B3MFX6 B4HRY2 B5DZA0 A0A0L0CMY3 A0A1I8ND58 B4KIU7 B4P2U4 A0A0M4EU81 A0A2A4K936 U5EJL5 B4P2T8 A0A3B0JKC9 A0A1L8EAF3 B4LLB2 A0A0L0CQC7 B4J5X3 A0A1L8EB26 A0A1L8EBI2 A0A1L8EB54 W8BNP2 B3N8P3 B4LLB7 O16099 A0A2W1BQ60 A0A1B0DP22 A0A2A4JCK8 Q29LL3 A0A0R3NTH9 B4KP52 B4GPS3 A0A0A1X1Q7 A0A212F7X9 P07190 B4KP54
A0A194PTX8 A0A194PPD0 A0A0D3R469 A0A0N0PB16 D8KY55 A0A2W1BJR6 A0A2A4JP22 A0A212F5X2 A0A0L7LPH9 A0A2W1BRC7 A0A2A4JNL4 H9J0L8 A0A2H1V8G1 A0A0N1INK8 A0A212F5W6 B4LLB9 B4KP53 Q292I4 W8ATZ7 W8AQ09 B4GDF4 A0A0N1I9T4 B4MQ20 A0A0A1XB46 A0A034WCA5 B4J5W7 A0A194QDR1 B3N8N6 A0A1I8N8F6 A0A3B0JRB9 B3MFX5 B4QFT0 A0A0J9R808 H5V882 A1Z7F3 A0A0M5J9Z6 A0A212F6K0 E0R939 B4J5X4 A0A1I8MGR6 B4KP60 T1PFH0 A0A3B0JJS3 A0A0L0CNA2 A0A1I8M319 B4P2U5 A0A1W4UXI6 A0A1W4UKB0 A0A212F7W3 B4MQ26 B4GDF3 B3MFY2 B3N8N7 A0A2H1W014 U5ELE7 A0A0M5IX79 A0A1I8PZX7 A0A0J9R8A6 A1Z7F2 A0A2A4JCN0 B3MFX6 B4HRY2 B5DZA0 A0A0L0CMY3 A0A1I8ND58 B4KIU7 B4P2U4 A0A0M4EU81 A0A2A4K936 U5EJL5 B4P2T8 A0A3B0JKC9 A0A1L8EAF3 B4LLB2 A0A0L0CQC7 B4J5X3 A0A1L8EB26 A0A1L8EBI2 A0A1L8EB54 W8BNP2 B3N8P3 B4LLB7 O16099 A0A2W1BQ60 A0A1B0DP22 A0A2A4JCK8 Q29LL3 A0A0R3NTH9 B4KP52 B4GPS3 A0A0A1X1Q7 A0A212F7X9 P07190 B4KP54
EC Number
3.2.1.20
Pubmed
EMBL
BABH01010222
NWSH01000913
PCG73560.1
AGBW02010122
OWR49125.1
KZ150000
+ More
PZC75321.1 ODYU01003827 SOQ43077.1 KQ461183 KPJ07576.1 KQ459598 KPI94590.1 KPI94589.1 KP101252 AJR29308.1 KPJ07577.1 EU856718 ACJ49024.1 PZC75322.1 PCG73559.1 OWR49124.1 JTDY01000407 KOB77337.1 PZC75320.1 PCG73561.1 BABH01010223 ODYU01001035 SOQ36692.1 KPJ07575.1 OWR49126.1 CH940648 EDW60856.1 CH933808 EDW10119.1 CM000071 EAL24878.2 GAMC01018417 JAB88138.1 GAMC01018418 JAB88137.1 CH479181 EDW31630.1 KQ460041 KPJ18359.1 CH963849 EDW74209.1 GBXI01005733 JAD08559.1 GAKP01007519 JAC51433.1 CH916367 EDW00810.1 KQ459324 KPJ01591.1 CH954177 EDV59513.1 OUUW01000001 SPP73688.1 CH902619 EDV35657.1 CM000362 EDX06186.1 CM002911 KMY92272.1 BT133183 AFA28424.1 AE013599 AAF59083.2 CP012524 ALC40931.1 AGBW02010006 OWR49357.1 BT125607 ADM26845.1 EDW00817.1 EDW10126.1 KA647537 AFP62166.1 SPP73689.1 JRES01000171 KNC33697.1 CM000157 EDW89356.1 AGBW02009824 OWR49826.1 EDW74215.1 EDW31629.1 EDV35664.1 EDV59514.1 ODYU01005517 SOQ46439.1 GANO01001436 JAB58435.1 ALC40924.1 KMY92271.1 AAF59084.2 AHN55996.1 NWSH01001872 PCG69835.1 EDV35658.1 CH480816 EDW46945.1 EDY68786.1 KNC33641.1 CH933807 EDW12453.2 EDW89355.1 ALC40932.1 NWSH01000047 PCG80230.1 GANO01002192 JAB57679.1 EDW89349.2 SPP73696.1 GFDG01003203 JAV15596.1 EDW60849.1 KNC33639.1 EDW00816.1 GFDG01002829 JAV15970.1 GFDG01002830 JAV15969.1 GFDG01002831 JAV15968.1 GAMC01003820 JAC02736.1 EDV59520.2 EDW60854.1 CH940649 AF006573 KZ149934 PZC77209.1 AJVK01007891 AJVK01007892 PCG69837.1 CH379060 EAL34032.3 KRT04418.1 EDW10118.2 CH479187 EDW39595.1 GBXI01009028 JAD05264.1 OWR49828.1 V00204 AY070626 EDW10120.1
PZC75321.1 ODYU01003827 SOQ43077.1 KQ461183 KPJ07576.1 KQ459598 KPI94590.1 KPI94589.1 KP101252 AJR29308.1 KPJ07577.1 EU856718 ACJ49024.1 PZC75322.1 PCG73559.1 OWR49124.1 JTDY01000407 KOB77337.1 PZC75320.1 PCG73561.1 BABH01010223 ODYU01001035 SOQ36692.1 KPJ07575.1 OWR49126.1 CH940648 EDW60856.1 CH933808 EDW10119.1 CM000071 EAL24878.2 GAMC01018417 JAB88138.1 GAMC01018418 JAB88137.1 CH479181 EDW31630.1 KQ460041 KPJ18359.1 CH963849 EDW74209.1 GBXI01005733 JAD08559.1 GAKP01007519 JAC51433.1 CH916367 EDW00810.1 KQ459324 KPJ01591.1 CH954177 EDV59513.1 OUUW01000001 SPP73688.1 CH902619 EDV35657.1 CM000362 EDX06186.1 CM002911 KMY92272.1 BT133183 AFA28424.1 AE013599 AAF59083.2 CP012524 ALC40931.1 AGBW02010006 OWR49357.1 BT125607 ADM26845.1 EDW00817.1 EDW10126.1 KA647537 AFP62166.1 SPP73689.1 JRES01000171 KNC33697.1 CM000157 EDW89356.1 AGBW02009824 OWR49826.1 EDW74215.1 EDW31629.1 EDV35664.1 EDV59514.1 ODYU01005517 SOQ46439.1 GANO01001436 JAB58435.1 ALC40924.1 KMY92271.1 AAF59084.2 AHN55996.1 NWSH01001872 PCG69835.1 EDV35658.1 CH480816 EDW46945.1 EDY68786.1 KNC33641.1 CH933807 EDW12453.2 EDW89355.1 ALC40932.1 NWSH01000047 PCG80230.1 GANO01002192 JAB57679.1 EDW89349.2 SPP73696.1 GFDG01003203 JAV15596.1 EDW60849.1 KNC33639.1 EDW00816.1 GFDG01002829 JAV15970.1 GFDG01002830 JAV15969.1 GFDG01002831 JAV15968.1 GAMC01003820 JAC02736.1 EDV59520.2 EDW60854.1 CH940649 AF006573 KZ149934 PZC77209.1 AJVK01007891 AJVK01007892 PCG69837.1 CH379060 EAL34032.3 KRT04418.1 EDW10118.2 CH479187 EDW39595.1 GBXI01009028 JAD05264.1 OWR49828.1 V00204 AY070626 EDW10120.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000008792 UP000009192 UP000001819 UP000008744 UP000007798 UP000001070 UP000008711 UP000095301 UP000268350 UP000007801 UP000000304 UP000000803 UP000092553 UP000037069 UP000002282 UP000192221 UP000095300 UP000001292 UP000092462
UP000008792 UP000009192 UP000001819 UP000008744 UP000007798 UP000001070 UP000008711 UP000095301 UP000268350 UP000007801 UP000000304 UP000000803 UP000092553 UP000037069 UP000002282 UP000192221 UP000095300 UP000001292 UP000092462
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
H9J0L9
A0A2A4JQ74
A0A212F5V9
A0A2W1BM09
A0A2H1VQK1
A0A0N0PAR2
+ More
A0A194PTX8 A0A194PPD0 A0A0D3R469 A0A0N0PB16 D8KY55 A0A2W1BJR6 A0A2A4JP22 A0A212F5X2 A0A0L7LPH9 A0A2W1BRC7 A0A2A4JNL4 H9J0L8 A0A2H1V8G1 A0A0N1INK8 A0A212F5W6 B4LLB9 B4KP53 Q292I4 W8ATZ7 W8AQ09 B4GDF4 A0A0N1I9T4 B4MQ20 A0A0A1XB46 A0A034WCA5 B4J5W7 A0A194QDR1 B3N8N6 A0A1I8N8F6 A0A3B0JRB9 B3MFX5 B4QFT0 A0A0J9R808 H5V882 A1Z7F3 A0A0M5J9Z6 A0A212F6K0 E0R939 B4J5X4 A0A1I8MGR6 B4KP60 T1PFH0 A0A3B0JJS3 A0A0L0CNA2 A0A1I8M319 B4P2U5 A0A1W4UXI6 A0A1W4UKB0 A0A212F7W3 B4MQ26 B4GDF3 B3MFY2 B3N8N7 A0A2H1W014 U5ELE7 A0A0M5IX79 A0A1I8PZX7 A0A0J9R8A6 A1Z7F2 A0A2A4JCN0 B3MFX6 B4HRY2 B5DZA0 A0A0L0CMY3 A0A1I8ND58 B4KIU7 B4P2U4 A0A0M4EU81 A0A2A4K936 U5EJL5 B4P2T8 A0A3B0JKC9 A0A1L8EAF3 B4LLB2 A0A0L0CQC7 B4J5X3 A0A1L8EB26 A0A1L8EBI2 A0A1L8EB54 W8BNP2 B3N8P3 B4LLB7 O16099 A0A2W1BQ60 A0A1B0DP22 A0A2A4JCK8 Q29LL3 A0A0R3NTH9 B4KP52 B4GPS3 A0A0A1X1Q7 A0A212F7X9 P07190 B4KP54
A0A194PTX8 A0A194PPD0 A0A0D3R469 A0A0N0PB16 D8KY55 A0A2W1BJR6 A0A2A4JP22 A0A212F5X2 A0A0L7LPH9 A0A2W1BRC7 A0A2A4JNL4 H9J0L8 A0A2H1V8G1 A0A0N1INK8 A0A212F5W6 B4LLB9 B4KP53 Q292I4 W8ATZ7 W8AQ09 B4GDF4 A0A0N1I9T4 B4MQ20 A0A0A1XB46 A0A034WCA5 B4J5W7 A0A194QDR1 B3N8N6 A0A1I8N8F6 A0A3B0JRB9 B3MFX5 B4QFT0 A0A0J9R808 H5V882 A1Z7F3 A0A0M5J9Z6 A0A212F6K0 E0R939 B4J5X4 A0A1I8MGR6 B4KP60 T1PFH0 A0A3B0JJS3 A0A0L0CNA2 A0A1I8M319 B4P2U5 A0A1W4UXI6 A0A1W4UKB0 A0A212F7W3 B4MQ26 B4GDF3 B3MFY2 B3N8N7 A0A2H1W014 U5ELE7 A0A0M5IX79 A0A1I8PZX7 A0A0J9R8A6 A1Z7F2 A0A2A4JCN0 B3MFX6 B4HRY2 B5DZA0 A0A0L0CMY3 A0A1I8ND58 B4KIU7 B4P2U4 A0A0M4EU81 A0A2A4K936 U5EJL5 B4P2T8 A0A3B0JKC9 A0A1L8EAF3 B4LLB2 A0A0L0CQC7 B4J5X3 A0A1L8EB26 A0A1L8EBI2 A0A1L8EB54 W8BNP2 B3N8P3 B4LLB7 O16099 A0A2W1BQ60 A0A1B0DP22 A0A2A4JCK8 Q29LL3 A0A0R3NTH9 B4KP52 B4GPS3 A0A0A1X1Q7 A0A212F7X9 P07190 B4KP54
PDB
3WY4
E-value=4.19808e-72,
Score=691
Ontologies
GO
Topology
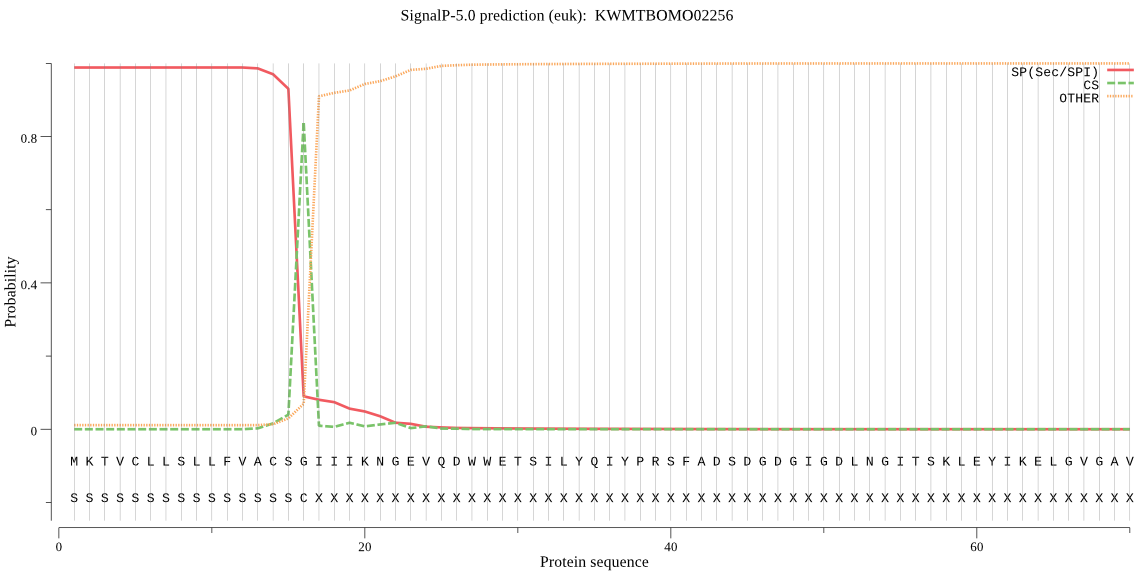
SignalP
Position: 1 - 16,
Likelihood: 0.988277
Length:
579
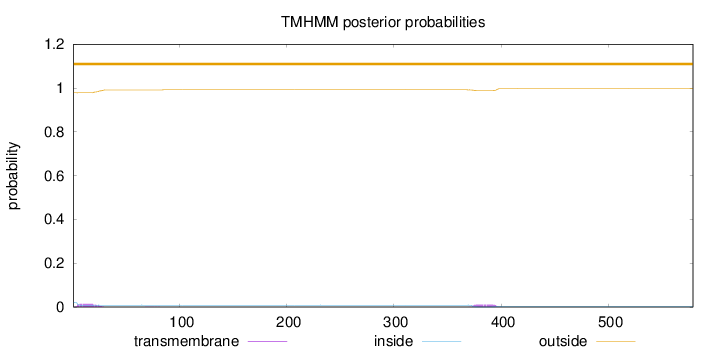
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.530179999999999
Exp number, first 60 AAs:
0.27966
Total prob of N-in:
0.02119
outside
1 - 579
Population Genetic Test Statistics
Pi
180.894055
Theta
179.191365
Tajima's D
-0.51084
CLR
1.725712
CSRT
0.23938803059847
Interpretation
Uncertain
