Gene
KWMTBOMO02253
Pre Gene Modal
BGIBMGA003099
Annotation
PREDICTED:_calcium/calmodulin-dependent_3'?5'-cyclic_nucleotide_phosphodiesterase_1A_isoform_X2_[Bombyx_mori]
Full name
Phosphodiesterase
+ More
Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1
Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1
Location in the cell
Cytoplasmic Reliability : 2.299 Nuclear Reliability : 1.788
Sequence
CDS
ATGGATGGTGCACTAACTTGTCCGGAGCAAGGCGCGCTGACAATCGTCCGCCGTTCTTCAAGCCGGAGTAAGACCGAGAGCTGCTCCACTCCTACTGACGAACAACTCGATCAGCTCGATCTAGAAACTGAACACTTGCCCGCCGTCGACACGCCCGACGCTTGCGATAAGGCCGCACTGAGATTACGCTATCTATTAAGACAATTAAACCGAGGTGAAATTTCTGCAGAGACTTTACAAAAGAACCTACAGTATGCTGCAAAAGTGCTTGAAGCTGTCTACATAGATGAGACCAAACGATTAGCTGATGAAGATGACGAGCTCTCTGAGGTACAGCCTGATGCGGTGCCCCCTGAGGTTCGCGAATGGCTGGCGTCGACATTCACAAGACAGCTGGCGACAGCCAAACGGAAGTCGGACGAGAAGCCAAAGTTCCGGTCGGTGGCCCATGCGATACGTGCCGGAATATTCGTCGACAGAATCTATCGCCGGGTGACCAGTACCGCTCTTATGCAGTTTCCTCAGGATGTCGTTAGAGTATTGAAGACGCTGGATGAATGGTCTTTCGACGTGTTCACTCTGCACGAGGCGGCCAACGGGCAACCGGTCAAATACTTAGGTTACGAACTGTTGAATCGATACGGAATGATTCATAAATTTAAGGTTCCTCCGACAATCTTAGAGAACTTCCTCGGCCGGATAGAGGAGGGCTACTGCAGGTTTCATAATCCCTACCACAATAACCTGCACGCCGCTGACGTCGCGCAAACCGTGCACTACATGCTGTGCCAGACCGGCCTTATGAACTGGCTCTCGGACCTGGAGATCTTCGCGACGCTGGTAGCCGCCGTGGTCCACGACTACGAGCACACGGGCACGACTAACAACTTCCACGTGATGTCCGGCTCGGACACCGCCCTGCTCTACAACGACAGGGCCGTGCTGGAGAACCATCACATCAGCGCCGCCTTCAGGTTAATGCGCGAAGAAGAATACAATATACTCCAGAATCTATCCCGGGATGAGTTTCGCGAATTTCGTACGCTTGTGATCGATATGGTGCTGGCGACCGACATGTCCTTCCACTTCCAGCAACTGAAGAACATGAGATCGCTTCTGTCCCTCGCAGAGCCAACCATAGACAAATCGAAGGCTACATCTCTGGTGCTCCATTGCTGCGACATTTCGCACCCTGCTAAGAGATGGGATCTCCACCACCGATGGACAATGAGTCTCTTGGATGAGTTTTTCCTCCAAGGTGACAAAGAACGAGATCTTGGTCTGCCCTTTAGTCCGCTCTGCGACAGAAATAATACTCTGGTGGCGGAATCCCAAATTGGTTTCATTGAGTTCATCGTGGAGCCCAGCATGGGAGTTTGTGCAGACATGCTGGAATGCATTTTGGGGCCTCTGCACTCCGGCAATAAATCCGGAACTAATGCAGCGACTGAACCGACTATTGATGAAAATTCGGGCGCCGAGAGTGTTAACAAATTCAAGATAAGGAAACCTTGGATTACCTGTCTCAGTGACAATAAGAAGATATGGAAGGAGCAAGCTACTAAAGACGCCGAGGCCCGAGCCAACCAGGAGCAGGAGGAGAACATAGAGGTTCAGCCTCCACCAGAAGAATAA
Protein
MDGALTCPEQGALTIVRRSSSRSKTESCSTPTDEQLDQLDLETEHLPAVDTPDACDKAALRLRYLLRQLNRGEISAETLQKNLQYAAKVLEAVYIDETKRLADEDDELSEVQPDAVPPEVREWLASTFTRQLATAKRKSDEKPKFRSVAHAIRAGIFVDRIYRRVTSTALMQFPQDVVRVLKTLDEWSFDVFTLHEAANGQPVKYLGYELLNRYGMIHKFKVPPTILENFLGRIEEGYCRFHNPYHNNLHAADVAQTVHYMLCQTGLMNWLSDLEIFATLVAAVVHDYEHTGTTNNFHVMSGSDTALLYNDRAVLENHHISAAFRLMREEEYNILQNLSRDEFREFRTLVIDMVLATDMSFHFQQLKNMRSLLSLAEPTIDKSKATSLVLHCCDISHPAKRWDLHHRWTMSLLDEFFLQGDKERDLGLPFSPLCDRNNTLVAESQIGFIEFIVEPSMGVCADMLECILGPLHSGNKSGTNAATEPTIDENSGAESVNKFKIRKPWITCLSDNKKIWKEQATKDAEARANQEQEENIEVQPPPEE
Summary
Description
Cyclic nucleotide phosphodiesterase with a dual specificity for the second messengers cAMP and cGMP, which are key regulators of many important physiological processes (PubMed:15673286). Required for male fertility and male mating behavior (PubMed:20551439).
Catalytic Activity
a nucleoside 3',5'-cyclic phosphate + H2O = a nucleoside 5'-phosphate + H(+)
Cofactor
a divalent metal cation
Biophysicochemical Properties
15.3 uM for cGMP
20.5 uM for cAMP
20.5 uM for cAMP
Similarity
Belongs to the cyclic nucleotide phosphodiesterase family.
Belongs to the cyclic nucleotide phosphodiesterase family. PDE1 subfamily.
Belongs to the cyclic nucleotide phosphodiesterase family. PDE1 subfamily.
Keywords
Alternative splicing
Behavior
cAMP
cGMP
Complete proteome
Hydrolase
Metal-binding
Reference proteome
Feature
chain Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1
splice variant In isoform C.
splice variant In isoform C.
Uniprot
H9J0R2
A0A194PMC9
A0A2W1BRM0
A0A151JXU5
A0A151HZH2
A0A151IDW8
+ More
E9IA42 A0A2A4J8S1 A0A154PFH1 A0A0L7RFY0 A0A151WS11 A0A146L145 A0A195EDA8 A0A1B6EBY1 K7IPZ8 A0A224XBF1 A0A1B6BYA4 A0A3L8D4P3 A0A0A9XIP6 A0A0A1WXK7 W8ASS9 A0A034VRC9 A0A067R622 A0A087ZWK2 E2A8S4 E2BBA7 A0A158NIF6 A0A310SH68 A0A026WWL0 A0A182LML7 A0A0Q9WA74 A0A0R3NU05 A0A084WQK0 A0A0R3P0H6 M9NF02 A0A0J9R105 A0A0J9R124 A0A0R1DRF1 A0A182QFA9 A0A0R1DK82 A0A182NUQ0 A0A0Q5WAP5 A0A0Q9WA77 A0A0P8XY04 A0A1B6KBN5 A0A0K8VVL3 A0A0R3NZG6 A0A182M968 B7YZV3 A0A1J1ILK9 A0A0J9R117 A0A0Q5WNW0 A0A1I8NIL7 A0A0P8XHM0 A0A182TDE4 A0A1I8PGH1 A0A0K8UGZ5 B4JR30 A0A0K8TWI6 W8B7P2 A0A0Q9WKH3 A0A0Q9WBF6 A0A0Q9WHC3 T1IN43 A0A0Q9WMH1 B4KIV6 A0A0Q9WA54 A0A0R3NTT5 A0A2S2N8D3 A0A0R3NTU7 A0A0R3NYR5 A0A0R3NTC2 A0A0R3NYG7 X1WIQ2 A0A0R3NTJ3 A0A0R3NT72 A0A0R3NZ09 B4GPR5 B4Q398 B7YZV4-2 B4MVY8 B4HX83 A0A0R3NTG6 B7YZV1 M9PDA0 A0A0R1DK37 A0A0J9R100 B7YZV2 A0A0J9TLL5 A0A0J9R0M6
E9IA42 A0A2A4J8S1 A0A154PFH1 A0A0L7RFY0 A0A151WS11 A0A146L145 A0A195EDA8 A0A1B6EBY1 K7IPZ8 A0A224XBF1 A0A1B6BYA4 A0A3L8D4P3 A0A0A9XIP6 A0A0A1WXK7 W8ASS9 A0A034VRC9 A0A067R622 A0A087ZWK2 E2A8S4 E2BBA7 A0A158NIF6 A0A310SH68 A0A026WWL0 A0A182LML7 A0A0Q9WA74 A0A0R3NU05 A0A084WQK0 A0A0R3P0H6 M9NF02 A0A0J9R105 A0A0J9R124 A0A0R1DRF1 A0A182QFA9 A0A0R1DK82 A0A182NUQ0 A0A0Q5WAP5 A0A0Q9WA77 A0A0P8XY04 A0A1B6KBN5 A0A0K8VVL3 A0A0R3NZG6 A0A182M968 B7YZV3 A0A1J1ILK9 A0A0J9R117 A0A0Q5WNW0 A0A1I8NIL7 A0A0P8XHM0 A0A182TDE4 A0A1I8PGH1 A0A0K8UGZ5 B4JR30 A0A0K8TWI6 W8B7P2 A0A0Q9WKH3 A0A0Q9WBF6 A0A0Q9WHC3 T1IN43 A0A0Q9WMH1 B4KIV6 A0A0Q9WA54 A0A0R3NTT5 A0A2S2N8D3 A0A0R3NTU7 A0A0R3NYR5 A0A0R3NTC2 A0A0R3NYG7 X1WIQ2 A0A0R3NTJ3 A0A0R3NT72 A0A0R3NZ09 B4GPR5 B4Q398 B7YZV4-2 B4MVY8 B4HX83 A0A0R3NTG6 B7YZV1 M9PDA0 A0A0R1DK37 A0A0J9R100 B7YZV2 A0A0J9TLL5 A0A0J9R0M6
EC Number
3.1.4.-
3.1.4.17
3.1.4.17
Pubmed
19121390
26354079
28756777
21282665
26823975
20075255
+ More
30249741 25401762 25830018 24495485 25348373 24845553 20798317 21347285 24508170 20966253 17994087 15632085 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 25315136 12537569 15673286 20551439
30249741 25401762 25830018 24495485 25348373 24845553 20798317 21347285 24508170 20966253 17994087 15632085 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 25315136 12537569 15673286 20551439
EMBL
BABH01010227
BABH01010228
BABH01010229
BABH01010230
KQ459598
KPI94591.1
+ More
KZ150000 PZC75316.1 KQ981607 KYN39385.1 KQ976695 KYM77576.1 KQ977939 KYM98535.1 GL761987 EFZ22551.1 NWSH01002627 PCG67812.1 KQ434893 KZC10585.1 KQ414599 KOC69743.1 KQ982796 KYQ50608.1 GDHC01017533 JAQ01096.1 KQ979074 KYN23081.1 GEDC01001856 JAS35442.1 GFTR01006680 JAW09746.1 GEDC01031031 JAS06267.1 QOIP01000014 RLU15111.1 GBHO01023022 JAG20582.1 GBXI01010690 JAD03602.1 GAMC01017315 GAMC01017313 GAMC01017307 JAB89242.1 GAKP01013101 JAC45851.1 KK852670 KDR18814.1 GL437663 EFN70153.1 GL446968 EFN87015.1 ADTU01016402 ADTU01016403 ADTU01016404 ADTU01016405 ADTU01016406 ADTU01016407 ADTU01016408 ADTU01016409 ADTU01016410 ADTU01016411 KQ769895 OAD52782.1 KK107083 EZA60096.1 CH940649 KRF81624.1 CH379060 KRT04412.1 ATLV01025637 KE525396 KFB52494.1 KRT04406.1 AE014134 AFH03668.1 CM002910 KMY89826.1 KMY89823.1 CM000157 KRJ97593.1 AXCN02000736 KRJ97592.1 CH954177 KQS70534.1 KRF81625.1 CH902624 KPU74341.1 GEBQ01031114 GEBQ01014034 JAT08863.1 JAT25943.1 GDHF01009400 JAI42914.1 KRT04416.1 AXCM01003796 ACL83025.2 CVRI01000054 CRL00642.1 KMY89818.1 KQS70536.1 KPU74342.1 GDHF01026375 JAI25939.1 CH916372 EDV99360.1 GDHF01033661 JAI18653.1 GAMC01017309 JAB89246.1 KRF81620.1 KRF81622.1 KRF81623.1 JH431140 KRF81621.1 CH933807 EDW12462.1 KRF81619.1 KRT04409.1 GGMR01000719 MBY13338.1 KRT04410.1 KRT04413.1 KRT04408.1 KRT04415.1 ABLF02037443 ABLF02037445 ABLF02037449 ABLF02037456 ABLF02037457 ABLF02037458 ABLF02037459 KRT04411.1 KRT04414.1 KRT04405.1 CH479187 EDW39587.1 CM000361 EDX04729.1 AY118396 AAM48425.1 CH963857 EDW75858.2 CH480818 EDW52628.1 KRT04407.1 ACL83024.1 AGB92930.1 KRJ97594.1 KMY89821.1 ACL83023.1 KMY89820.1 KMY89822.1
KZ150000 PZC75316.1 KQ981607 KYN39385.1 KQ976695 KYM77576.1 KQ977939 KYM98535.1 GL761987 EFZ22551.1 NWSH01002627 PCG67812.1 KQ434893 KZC10585.1 KQ414599 KOC69743.1 KQ982796 KYQ50608.1 GDHC01017533 JAQ01096.1 KQ979074 KYN23081.1 GEDC01001856 JAS35442.1 GFTR01006680 JAW09746.1 GEDC01031031 JAS06267.1 QOIP01000014 RLU15111.1 GBHO01023022 JAG20582.1 GBXI01010690 JAD03602.1 GAMC01017315 GAMC01017313 GAMC01017307 JAB89242.1 GAKP01013101 JAC45851.1 KK852670 KDR18814.1 GL437663 EFN70153.1 GL446968 EFN87015.1 ADTU01016402 ADTU01016403 ADTU01016404 ADTU01016405 ADTU01016406 ADTU01016407 ADTU01016408 ADTU01016409 ADTU01016410 ADTU01016411 KQ769895 OAD52782.1 KK107083 EZA60096.1 CH940649 KRF81624.1 CH379060 KRT04412.1 ATLV01025637 KE525396 KFB52494.1 KRT04406.1 AE014134 AFH03668.1 CM002910 KMY89826.1 KMY89823.1 CM000157 KRJ97593.1 AXCN02000736 KRJ97592.1 CH954177 KQS70534.1 KRF81625.1 CH902624 KPU74341.1 GEBQ01031114 GEBQ01014034 JAT08863.1 JAT25943.1 GDHF01009400 JAI42914.1 KRT04416.1 AXCM01003796 ACL83025.2 CVRI01000054 CRL00642.1 KMY89818.1 KQS70536.1 KPU74342.1 GDHF01026375 JAI25939.1 CH916372 EDV99360.1 GDHF01033661 JAI18653.1 GAMC01017309 JAB89246.1 KRF81620.1 KRF81622.1 KRF81623.1 JH431140 KRF81621.1 CH933807 EDW12462.1 KRF81619.1 KRT04409.1 GGMR01000719 MBY13338.1 KRT04410.1 KRT04413.1 KRT04408.1 KRT04415.1 ABLF02037443 ABLF02037445 ABLF02037449 ABLF02037456 ABLF02037457 ABLF02037458 ABLF02037459 KRT04411.1 KRT04414.1 KRT04405.1 CH479187 EDW39587.1 CM000361 EDX04729.1 AY118396 AAM48425.1 CH963857 EDW75858.2 CH480818 EDW52628.1 KRT04407.1 ACL83024.1 AGB92930.1 KRJ97594.1 KMY89821.1 ACL83023.1 KMY89820.1 KMY89822.1
Proteomes
UP000005204
UP000053268
UP000078541
UP000078540
UP000078542
UP000218220
+ More
UP000076502 UP000053825 UP000075809 UP000078492 UP000002358 UP000279307 UP000027135 UP000005203 UP000000311 UP000008237 UP000005205 UP000053097 UP000075882 UP000008792 UP000001819 UP000030765 UP000000803 UP000002282 UP000075886 UP000075884 UP000008711 UP000007801 UP000075883 UP000183832 UP000095301 UP000075902 UP000095300 UP000001070 UP000009192 UP000007819 UP000008744 UP000000304 UP000007798 UP000001292
UP000076502 UP000053825 UP000075809 UP000078492 UP000002358 UP000279307 UP000027135 UP000005203 UP000000311 UP000008237 UP000005205 UP000053097 UP000075882 UP000008792 UP000001819 UP000030765 UP000000803 UP000002282 UP000075886 UP000075884 UP000008711 UP000007801 UP000075883 UP000183832 UP000095301 UP000075902 UP000095300 UP000001070 UP000009192 UP000007819 UP000008744 UP000000304 UP000007798 UP000001292
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J0R2
A0A194PMC9
A0A2W1BRM0
A0A151JXU5
A0A151HZH2
A0A151IDW8
+ More
E9IA42 A0A2A4J8S1 A0A154PFH1 A0A0L7RFY0 A0A151WS11 A0A146L145 A0A195EDA8 A0A1B6EBY1 K7IPZ8 A0A224XBF1 A0A1B6BYA4 A0A3L8D4P3 A0A0A9XIP6 A0A0A1WXK7 W8ASS9 A0A034VRC9 A0A067R622 A0A087ZWK2 E2A8S4 E2BBA7 A0A158NIF6 A0A310SH68 A0A026WWL0 A0A182LML7 A0A0Q9WA74 A0A0R3NU05 A0A084WQK0 A0A0R3P0H6 M9NF02 A0A0J9R105 A0A0J9R124 A0A0R1DRF1 A0A182QFA9 A0A0R1DK82 A0A182NUQ0 A0A0Q5WAP5 A0A0Q9WA77 A0A0P8XY04 A0A1B6KBN5 A0A0K8VVL3 A0A0R3NZG6 A0A182M968 B7YZV3 A0A1J1ILK9 A0A0J9R117 A0A0Q5WNW0 A0A1I8NIL7 A0A0P8XHM0 A0A182TDE4 A0A1I8PGH1 A0A0K8UGZ5 B4JR30 A0A0K8TWI6 W8B7P2 A0A0Q9WKH3 A0A0Q9WBF6 A0A0Q9WHC3 T1IN43 A0A0Q9WMH1 B4KIV6 A0A0Q9WA54 A0A0R3NTT5 A0A2S2N8D3 A0A0R3NTU7 A0A0R3NYR5 A0A0R3NTC2 A0A0R3NYG7 X1WIQ2 A0A0R3NTJ3 A0A0R3NT72 A0A0R3NZ09 B4GPR5 B4Q398 B7YZV4-2 B4MVY8 B4HX83 A0A0R3NTG6 B7YZV1 M9PDA0 A0A0R1DK37 A0A0J9R100 B7YZV2 A0A0J9TLL5 A0A0J9R0M6
E9IA42 A0A2A4J8S1 A0A154PFH1 A0A0L7RFY0 A0A151WS11 A0A146L145 A0A195EDA8 A0A1B6EBY1 K7IPZ8 A0A224XBF1 A0A1B6BYA4 A0A3L8D4P3 A0A0A9XIP6 A0A0A1WXK7 W8ASS9 A0A034VRC9 A0A067R622 A0A087ZWK2 E2A8S4 E2BBA7 A0A158NIF6 A0A310SH68 A0A026WWL0 A0A182LML7 A0A0Q9WA74 A0A0R3NU05 A0A084WQK0 A0A0R3P0H6 M9NF02 A0A0J9R105 A0A0J9R124 A0A0R1DRF1 A0A182QFA9 A0A0R1DK82 A0A182NUQ0 A0A0Q5WAP5 A0A0Q9WA77 A0A0P8XY04 A0A1B6KBN5 A0A0K8VVL3 A0A0R3NZG6 A0A182M968 B7YZV3 A0A1J1ILK9 A0A0J9R117 A0A0Q5WNW0 A0A1I8NIL7 A0A0P8XHM0 A0A182TDE4 A0A1I8PGH1 A0A0K8UGZ5 B4JR30 A0A0K8TWI6 W8B7P2 A0A0Q9WKH3 A0A0Q9WBF6 A0A0Q9WHC3 T1IN43 A0A0Q9WMH1 B4KIV6 A0A0Q9WA54 A0A0R3NTT5 A0A2S2N8D3 A0A0R3NTU7 A0A0R3NYR5 A0A0R3NTC2 A0A0R3NYG7 X1WIQ2 A0A0R3NTJ3 A0A0R3NT72 A0A0R3NZ09 B4GPR5 B4Q398 B7YZV4-2 B4MVY8 B4HX83 A0A0R3NTG6 B7YZV1 M9PDA0 A0A0R1DK37 A0A0J9R100 B7YZV2 A0A0J9TLL5 A0A0J9R0M6
PDB
5W6E
E-value=1.03048e-104,
Score=972
Ontologies
PATHWAY
GO
Topology
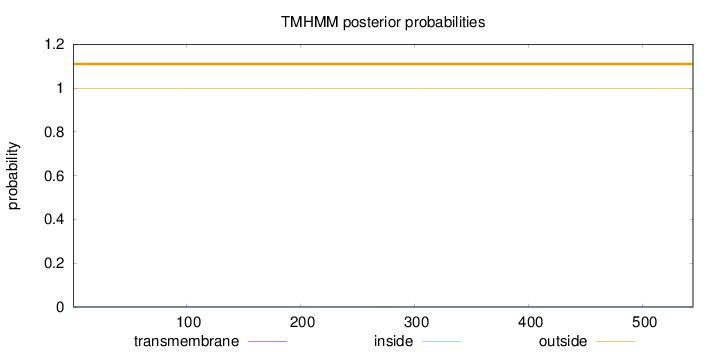
Length:
544
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00549
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00087
outside
1 - 544
Population Genetic Test Statistics
Pi
249.668062
Theta
169.250427
Tajima's D
1.189844
CLR
1.096175
CSRT
0.719914004299785
Interpretation
Uncertain
