Pre Gene Modal
BGIBMGA003053
Annotation
Arginine_methyltransferase_3_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 2.005 Nuclear Reliability : 1.658
Sequence
CDS
ATGATAAACTACATAAAGACAAACAAGCCAACTGCTGAAGAATTTATGAAAATTAAGCACCCGCCTTGGAACGACGACAAGTATTTGAAACCTGTTGAGAATGACGAATGGCTCACATTCGACTTCGATTCTGTATCTGAAAAACCTAATTCCCCCAAAACATATCATGCCAATGTTGAGAATGGTGTTGTTACTCTTTCTCAGGGACATTTTACTGAATTACAAAAAACCATACAAACATTGAGCGAACAGTTAAGTGAGAGTCAAACTCATCTGCAAATGGCCAAGGAGGATATAAACAAAATGCAAGACTCAATGAAAACATTAGTTGAAGGCGGCCAGTCCTCAAGTTGTGAAGTAAAAAATAAAACAAGTTGTGTTGCGAGTATTCCTATAGAGTGTGACGAAGGATACTTTAACACTTATGCACACTTTGGAATTCATTACGATATGCTCTCAGATAAAGTACGAACAGAAACGTATCGTGATGCCATATTAAAAAATAAAGAAACCTTAAGAGAGAAGACTGTACTTGATCTTGGTTGTGGCACAGGAATACTGTCAATGTTTGCTGCATTAGCAGGTGCCAAACAAATTTATGCCTTAGACCAATCTGATATAATATACCATGCAATGGATATAATAACTGAAAACAATTTACAAAATGTAATAAAGACAATTAAAGGTAGACTTGAAGAAACAAAACTTGAAGAAAAAGTTGATGTATTAGTTTCCGAGTGGATGGGATACTTCTTATTATTTGAAGGAATGTTGGATAGTGTAATATATGCCAGAGATAACTGTTTGAAGTCAGGTGGTTTGTTAATGCCTAATAGATGTAGCATAAGTATTGTAGCAAATGGTGATGTTGATACACACAAGAAGCTAATAGATTTTTGGTCTGATGTTTATGGTTTTAAAATGAACTGTATGAAATCTGAAGTAGTAAGAGAGGCCAGTATAGACGTAGTTGCATCTAAATATATATTGTCTCAACCATATGTTATAAATGAAATAGACATTAGTACTTGTGATACTACCGTAATGGATTTTACATCAGAATTTAAATTGGAGATTCTAAGGGATGGTTATTTGACATCTTTAGTAGGATATTTTGATACGTTTTTTGATCTGCCCAATAGTGTGTCATTTTCAACAGGTCCTCATTCAGCACCTACACACTGGAAACAAACTGTATTTTATTTAAGAGATTGTAAAGAGGTTAAACAAGGCAGTGTTATAACTGGCACCATAATTTGTAATAGGCAAAAAACAGATGTCCGAGCATTATCTGTGCAGATAGAGATATTTGGAAAAAATCACAAATATATACTAAGTTAG
Protein
MINYIKTNKPTAEEFMKIKHPPWNDDKYLKPVENDEWLTFDFDSVSEKPNSPKTYHANVENGVVTLSQGHFTELQKTIQTLSEQLSESQTHLQMAKEDINKMQDSMKTLVEGGQSSSCEVKNKTSCVASIPIECDEGYFNTYAHFGIHYDMLSDKVRTETYRDAILKNKETLREKTVLDLGCGTGILSMFAALAGAKQIYALDQSDIIYHAMDIITENNLQNVIKTIKGRLEETKLEEKVDVLVSEWMGYFLLFEGMLDSVIYARDNCLKSGGLLMPNRCSISIVANGDVDTHKKLIDFWSDVYGFKMNCMKSEVVREASIDVVASKYILSQPYVINEIDISTCDTTVMDFTSEFKLEILRDGYLTSLVGYFDTFFDLPNSVSFSTGPHSAPTHWKQTVFYLRDCKEVKQGSVITGTIICNRQKTDVRALSVQIEIFGKNHKYILS
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. Protein arginine N-methyltransferase family.
Uniprot
H9J0L6
A0A0L7LR59
A0A2H1VLU3
A0A2W1BNX5
A0A2A4J8U3
A0A212FKP0
+ More
A0A194PMQ0 A0A0N1I4Y3 S4P8I8 A0A2J7QS96 A0A2J7QS66 A0A067RDQ9 A0A1W4WVW7 A0A2J7QS71 A0A1W4WUV2 A0A1B6EFJ4 A0A1B6C1N1 A0A0C9R400 A0A0C9RGN1 A0A182QT65 A0A2M4BJ53 A0A1Q3F1J6 A0A182FNS4 B0WQZ2 W5JJ92 A0A1B6LVQ2 A0A1Y9HDI2 A0A182LZP7 A0A182JAL2 A0A2M3Z5V2 A0A1Y9IVG2 A0A2M4AJ14 A0A2M4AJ80 A0A2J7QS72 A0A232F268 K7J2R6 A0A2J7QS74 A0A1B6JFP7 A0A182N4R1 A0A182GRN0 A0A182WVR5 A0A088A326 A0A182JRJ4 A0A3F2YWI5 A0A2A3E8W7 A0A182V917 A0A182Y7C3 A0A1Y9GLE8 A0A154PEH6 A0A2M4AKY1 Q7QAP5 A0A0N0U3Z3 J3JUA3 A0A0T6AWZ2 A0A0L7QQX0 A0A084VHA0 N6UPF6 A0A182TDX4 D1ZZP4 U5EY54 E2BUE7 A0A182KXJ8 A0A310SAW5 A0A026VSQ2 Q0IED7 E0VBG9 W8CB50 A0A1J1I011 A0A1I8MMA5 W8BQS3 A0A1B0CVD2 B3P0S2 A0A0L0BZY4 A0A034WT15 B4HE04 A0A195ETE7 A0A0K8U7R6 A0A1I8P4M4 A0A158NPL1 A0A1B3PD56 F4W7P2 Q8INE8 A0A0A1WYI9 A0A1L8DMT1 Q9VFB3 A0A2P8Y071 B4PS75 B3MX97 A0A0P8ZL65 A0A336MBG4 A0A0P5DEH3 A0A0P5EFF0 A0A1B0D2Y6 A0A1W4W116 A0A0N8DW14 A0A1S4FRR7 T1E9J6 A0A0P6IE84 A0A165AAR5 A0A1B3PDH0
A0A194PMQ0 A0A0N1I4Y3 S4P8I8 A0A2J7QS96 A0A2J7QS66 A0A067RDQ9 A0A1W4WVW7 A0A2J7QS71 A0A1W4WUV2 A0A1B6EFJ4 A0A1B6C1N1 A0A0C9R400 A0A0C9RGN1 A0A182QT65 A0A2M4BJ53 A0A1Q3F1J6 A0A182FNS4 B0WQZ2 W5JJ92 A0A1B6LVQ2 A0A1Y9HDI2 A0A182LZP7 A0A182JAL2 A0A2M3Z5V2 A0A1Y9IVG2 A0A2M4AJ14 A0A2M4AJ80 A0A2J7QS72 A0A232F268 K7J2R6 A0A2J7QS74 A0A1B6JFP7 A0A182N4R1 A0A182GRN0 A0A182WVR5 A0A088A326 A0A182JRJ4 A0A3F2YWI5 A0A2A3E8W7 A0A182V917 A0A182Y7C3 A0A1Y9GLE8 A0A154PEH6 A0A2M4AKY1 Q7QAP5 A0A0N0U3Z3 J3JUA3 A0A0T6AWZ2 A0A0L7QQX0 A0A084VHA0 N6UPF6 A0A182TDX4 D1ZZP4 U5EY54 E2BUE7 A0A182KXJ8 A0A310SAW5 A0A026VSQ2 Q0IED7 E0VBG9 W8CB50 A0A1J1I011 A0A1I8MMA5 W8BQS3 A0A1B0CVD2 B3P0S2 A0A0L0BZY4 A0A034WT15 B4HE04 A0A195ETE7 A0A0K8U7R6 A0A1I8P4M4 A0A158NPL1 A0A1B3PD56 F4W7P2 Q8INE8 A0A0A1WYI9 A0A1L8DMT1 Q9VFB3 A0A2P8Y071 B4PS75 B3MX97 A0A0P8ZL65 A0A336MBG4 A0A0P5DEH3 A0A0P5EFF0 A0A1B0D2Y6 A0A1W4W116 A0A0N8DW14 A0A1S4FRR7 T1E9J6 A0A0P6IE84 A0A165AAR5 A0A1B3PDH0
Pubmed
19121390
26227816
28756777
22118469
26354079
23622113
+ More
24845553 20920257 23761445 28648823 20075255 26483478 25244985 12364791 14747013 17210077 22516182 23537049 24438588 18362917 19820115 20798317 20966253 24508170 17510324 20566863 24495485 25315136 17994087 26108605 25348373 21347285 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 29403074 17550304
24845553 20920257 23761445 28648823 20075255 26483478 25244985 12364791 14747013 17210077 22516182 23537049 24438588 18362917 19820115 20798317 20966253 24508170 17510324 20566863 24495485 25315136 17994087 26108605 25348373 21347285 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 29403074 17550304
EMBL
BABH01010231
JTDY01000270
KOB77988.1
ODYU01003270
SOQ41805.1
KZ150000
+ More
PZC75315.1 NWSH01002627 PCG67813.1 AGBW02008010 OWR54302.1 KQ459598 KPI94592.1 KQ461183 KPJ07572.1 GAIX01004184 JAA88376.1 NEVH01011878 PNF31443.1 PNF31441.1 KK852761 KDR16996.1 PNF31442.1 GEDC01000638 JAS36660.1 GEDC01030109 JAS07189.1 GBYB01007532 JAG77299.1 GBYB01007530 GBYB01007533 JAG77297.1 JAG77300.1 AXCN02000015 GGFJ01003921 MBW53062.1 GFDL01013612 JAV21433.1 DS232047 EDS33079.1 ADMH02001019 ETN64427.1 GEBQ01012224 JAT27753.1 AXCM01001019 GGFM01003145 MBW23896.1 GGFK01007453 MBW40774.1 GGFK01007510 MBW40831.1 PNF31439.1 NNAY01001258 OXU24593.1 PNF31438.1 GECU01009707 JAS97999.1 JXUM01082819 KQ563338 KXJ74021.1 KZ288322 PBC28167.1 APCN01005540 KQ434889 KZC10241.1 GGFK01008124 MBW41445.1 AAAB01008888 EAA08812.5 KQ435878 KOX70074.1 BT126816 KB632308 AEE61778.1 ERL91994.1 LJIG01022617 KRT79606.1 KQ414786 KOC60886.1 ATLV01013150 KE524842 KFB37344.1 APGK01023184 KB740473 ENN80587.1 KQ971338 EFA02398.1 GANO01000520 JAB59351.1 GL450640 EFN80674.1 KQ765605 OAD53841.1 KK108267 EZA46803.1 CH477695 EAT37162.1 DS235030 EEB10725.1 GAMC01005141 JAC01415.1 CVRI01000024 CRK92142.1 GAMC01005143 JAC01413.1 AJWK01030434 CH954181 EDV48970.1 JRES01001097 KNC25546.1 GAKP01002029 JAC56923.1 CH480815 EDW42096.1 KQ981986 KYN31182.1 GDHF01029934 JAI22380.1 ADTU01022588 KU659865 AOG17664.1 GL887844 EGI69915.1 AE014297 BT003167 AAN13635.1 AAO24922.1 GBXI01010576 JAD03716.1 GFDF01006434 JAV07650.1 AY051841 AAF55147.1 AAK93265.1 PYGN01001092 PSN37657.1 CM000160 EDW97495.1 CH902629 EDV35320.1 KPU75546.1 UFQT01000611 SSX25717.1 GDIP01157588 JAJ65814.1 GDIP01144030 JAJ79372.1 AJVK01023141 GDIQ01086356 JAN08381.1 GAMD01001793 JAA99797.1 GDIQ01005822 JAN88915.1 LRGB01000642 KZS17383.1 KU659975 AOG17773.1
PZC75315.1 NWSH01002627 PCG67813.1 AGBW02008010 OWR54302.1 KQ459598 KPI94592.1 KQ461183 KPJ07572.1 GAIX01004184 JAA88376.1 NEVH01011878 PNF31443.1 PNF31441.1 KK852761 KDR16996.1 PNF31442.1 GEDC01000638 JAS36660.1 GEDC01030109 JAS07189.1 GBYB01007532 JAG77299.1 GBYB01007530 GBYB01007533 JAG77297.1 JAG77300.1 AXCN02000015 GGFJ01003921 MBW53062.1 GFDL01013612 JAV21433.1 DS232047 EDS33079.1 ADMH02001019 ETN64427.1 GEBQ01012224 JAT27753.1 AXCM01001019 GGFM01003145 MBW23896.1 GGFK01007453 MBW40774.1 GGFK01007510 MBW40831.1 PNF31439.1 NNAY01001258 OXU24593.1 PNF31438.1 GECU01009707 JAS97999.1 JXUM01082819 KQ563338 KXJ74021.1 KZ288322 PBC28167.1 APCN01005540 KQ434889 KZC10241.1 GGFK01008124 MBW41445.1 AAAB01008888 EAA08812.5 KQ435878 KOX70074.1 BT126816 KB632308 AEE61778.1 ERL91994.1 LJIG01022617 KRT79606.1 KQ414786 KOC60886.1 ATLV01013150 KE524842 KFB37344.1 APGK01023184 KB740473 ENN80587.1 KQ971338 EFA02398.1 GANO01000520 JAB59351.1 GL450640 EFN80674.1 KQ765605 OAD53841.1 KK108267 EZA46803.1 CH477695 EAT37162.1 DS235030 EEB10725.1 GAMC01005141 JAC01415.1 CVRI01000024 CRK92142.1 GAMC01005143 JAC01413.1 AJWK01030434 CH954181 EDV48970.1 JRES01001097 KNC25546.1 GAKP01002029 JAC56923.1 CH480815 EDW42096.1 KQ981986 KYN31182.1 GDHF01029934 JAI22380.1 ADTU01022588 KU659865 AOG17664.1 GL887844 EGI69915.1 AE014297 BT003167 AAN13635.1 AAO24922.1 GBXI01010576 JAD03716.1 GFDF01006434 JAV07650.1 AY051841 AAF55147.1 AAK93265.1 PYGN01001092 PSN37657.1 CM000160 EDW97495.1 CH902629 EDV35320.1 KPU75546.1 UFQT01000611 SSX25717.1 GDIP01157588 JAJ65814.1 GDIP01144030 JAJ79372.1 AJVK01023141 GDIQ01086356 JAN08381.1 GAMD01001793 JAA99797.1 GDIQ01005822 JAN88915.1 LRGB01000642 KZS17383.1 KU659975 AOG17773.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000235965 UP000027135 UP000192223 UP000075886 UP000069272 UP000002320 UP000000673 UP000075900 UP000075883 UP000075880 UP000075920 UP000215335 UP000002358 UP000075884 UP000069940 UP000249989 UP000076407 UP000005203 UP000075881 UP000075885 UP000242457 UP000075903 UP000076408 UP000075840 UP000076502 UP000007062 UP000053105 UP000030742 UP000053825 UP000030765 UP000019118 UP000075902 UP000007266 UP000008237 UP000075882 UP000053097 UP000008820 UP000009046 UP000183832 UP000095301 UP000092461 UP000008711 UP000037069 UP000001292 UP000078541 UP000095300 UP000005205 UP000007755 UP000000803 UP000245037 UP000002282 UP000007801 UP000092462 UP000192221 UP000076858
UP000235965 UP000027135 UP000192223 UP000075886 UP000069272 UP000002320 UP000000673 UP000075900 UP000075883 UP000075880 UP000075920 UP000215335 UP000002358 UP000075884 UP000069940 UP000249989 UP000076407 UP000005203 UP000075881 UP000075885 UP000242457 UP000075903 UP000076408 UP000075840 UP000076502 UP000007062 UP000053105 UP000030742 UP000053825 UP000030765 UP000019118 UP000075902 UP000007266 UP000008237 UP000075882 UP000053097 UP000008820 UP000009046 UP000183832 UP000095301 UP000092461 UP000008711 UP000037069 UP000001292 UP000078541 UP000095300 UP000005205 UP000007755 UP000000803 UP000245037 UP000002282 UP000007801 UP000092462 UP000192221 UP000076858
Pfam
Interpro
ProteinModelPortal
H9J0L6
A0A0L7LR59
A0A2H1VLU3
A0A2W1BNX5
A0A2A4J8U3
A0A212FKP0
+ More
A0A194PMQ0 A0A0N1I4Y3 S4P8I8 A0A2J7QS96 A0A2J7QS66 A0A067RDQ9 A0A1W4WVW7 A0A2J7QS71 A0A1W4WUV2 A0A1B6EFJ4 A0A1B6C1N1 A0A0C9R400 A0A0C9RGN1 A0A182QT65 A0A2M4BJ53 A0A1Q3F1J6 A0A182FNS4 B0WQZ2 W5JJ92 A0A1B6LVQ2 A0A1Y9HDI2 A0A182LZP7 A0A182JAL2 A0A2M3Z5V2 A0A1Y9IVG2 A0A2M4AJ14 A0A2M4AJ80 A0A2J7QS72 A0A232F268 K7J2R6 A0A2J7QS74 A0A1B6JFP7 A0A182N4R1 A0A182GRN0 A0A182WVR5 A0A088A326 A0A182JRJ4 A0A3F2YWI5 A0A2A3E8W7 A0A182V917 A0A182Y7C3 A0A1Y9GLE8 A0A154PEH6 A0A2M4AKY1 Q7QAP5 A0A0N0U3Z3 J3JUA3 A0A0T6AWZ2 A0A0L7QQX0 A0A084VHA0 N6UPF6 A0A182TDX4 D1ZZP4 U5EY54 E2BUE7 A0A182KXJ8 A0A310SAW5 A0A026VSQ2 Q0IED7 E0VBG9 W8CB50 A0A1J1I011 A0A1I8MMA5 W8BQS3 A0A1B0CVD2 B3P0S2 A0A0L0BZY4 A0A034WT15 B4HE04 A0A195ETE7 A0A0K8U7R6 A0A1I8P4M4 A0A158NPL1 A0A1B3PD56 F4W7P2 Q8INE8 A0A0A1WYI9 A0A1L8DMT1 Q9VFB3 A0A2P8Y071 B4PS75 B3MX97 A0A0P8ZL65 A0A336MBG4 A0A0P5DEH3 A0A0P5EFF0 A0A1B0D2Y6 A0A1W4W116 A0A0N8DW14 A0A1S4FRR7 T1E9J6 A0A0P6IE84 A0A165AAR5 A0A1B3PDH0
A0A194PMQ0 A0A0N1I4Y3 S4P8I8 A0A2J7QS96 A0A2J7QS66 A0A067RDQ9 A0A1W4WVW7 A0A2J7QS71 A0A1W4WUV2 A0A1B6EFJ4 A0A1B6C1N1 A0A0C9R400 A0A0C9RGN1 A0A182QT65 A0A2M4BJ53 A0A1Q3F1J6 A0A182FNS4 B0WQZ2 W5JJ92 A0A1B6LVQ2 A0A1Y9HDI2 A0A182LZP7 A0A182JAL2 A0A2M3Z5V2 A0A1Y9IVG2 A0A2M4AJ14 A0A2M4AJ80 A0A2J7QS72 A0A232F268 K7J2R6 A0A2J7QS74 A0A1B6JFP7 A0A182N4R1 A0A182GRN0 A0A182WVR5 A0A088A326 A0A182JRJ4 A0A3F2YWI5 A0A2A3E8W7 A0A182V917 A0A182Y7C3 A0A1Y9GLE8 A0A154PEH6 A0A2M4AKY1 Q7QAP5 A0A0N0U3Z3 J3JUA3 A0A0T6AWZ2 A0A0L7QQX0 A0A084VHA0 N6UPF6 A0A182TDX4 D1ZZP4 U5EY54 E2BUE7 A0A182KXJ8 A0A310SAW5 A0A026VSQ2 Q0IED7 E0VBG9 W8CB50 A0A1J1I011 A0A1I8MMA5 W8BQS3 A0A1B0CVD2 B3P0S2 A0A0L0BZY4 A0A034WT15 B4HE04 A0A195ETE7 A0A0K8U7R6 A0A1I8P4M4 A0A158NPL1 A0A1B3PD56 F4W7P2 Q8INE8 A0A0A1WYI9 A0A1L8DMT1 Q9VFB3 A0A2P8Y071 B4PS75 B3MX97 A0A0P8ZL65 A0A336MBG4 A0A0P5DEH3 A0A0P5EFF0 A0A1B0D2Y6 A0A1W4W116 A0A0N8DW14 A0A1S4FRR7 T1E9J6 A0A0P6IE84 A0A165AAR5 A0A1B3PDH0
PDB
4RYL
E-value=8.73178e-87,
Score=817
Ontologies
GO
Topology
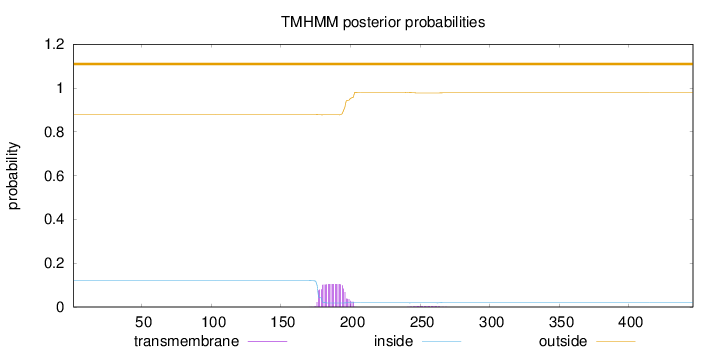
Length:
446
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.23721
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12077
outside
1 - 446
Population Genetic Test Statistics
Pi
189.45362
Theta
225.638128
Tajima's D
-0.505002
CLR
0.307584
CSRT
0.243537823108845
Interpretation
Uncertain
