Gene
KWMTBOMO02249
Pre Gene Modal
BGIBMGA003052
Annotation
PREDICTED:_acid_trehalase-like_protein_1_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.143 Peroxisomal Reliability : 1.035
Sequence
CDS
ATGTGGCGATTTTTGACGCGAAATAAACAAGTGGCGGCCATGATGGGCTTACTCACCGTCGCCGCTTTGGTCGTCGTTGTAATGACAACTAAAGGTAGCGAAGGTGGCCATGCGGCTGCTAATGATGAAGAGCCTGAGGATATCAACAACGATCCTAAAGTTTTTTCCACATTCAGGTTGCCAACCAACGATAGATTCATGGCGTCTATAGGCAATGGTCACGTAGCAGCGAACGTATTCAGCGATACGGTCTACATGAACGGATTGTATAACGGGAACAAAGGCGAGAGTCATCGTGCTAGGATACCTAACTGGGCCAACATCAGACTGAATTCGACCTTATCGCACATCCCGTACAGTCCAGTTTACTCTTTAGATACAAAAGAAGGCGCCTTTAAAGTCAGAGTTGATAGAGAGAGGTCGATTGTGACGCAGAGGATATTCGCTCATCGGTATTACACGAGGGCTATTATTAATCAGATCCAAGTTATGGCTAAAGCGCACGCCGAACCAAATTTTGTTCGCCACGAGATATGGATTGCGATACAATTAATTCCTGGACCGGATAGCGAGGATATCGGCTTCAAGGATCCCGTTCCCGAGATAATTGAAGGACGCACAGTCCGGAGTACGTGTGGTCAAACTTTGAAATCGGAGGATATCGTATATCAGCCACTGCCGGTGGAGGTCTGCGTGTTCTGGACGTCGGTGCCTGATCACTTGGTGGCCCCGCAGCACGGGTCCAGGGTTTTCACGTTTGTTATGACAGCGGATACCAATAGGACAGTTGCCAGAGAGGAGATGATCAGAGTTCTACAAGAAGACGGAGAAGAGCTTTACGATAAGCACGTGGCCGAGTGGCAGAAGCTTCATCAGCAAGCTCACATTGAAATTGAAGGGAATTTGAAAGTTGCAAAGGTTGTAAATGCAATCTGGTACTACTTCCTCAGTTCGTTGCCTTCGGAGGATTCACATCAGCCATTAGGCAGGTTTTACGGCCTCAGCCCAACAGGACTCGCCAGAGGAGGGACTTTGGAAGATTACGAAGGTCACAATTTCTGGGACACCGAAATCTGGATGTTTCCGTCTATTTTACTTCTATATCCGAAGTATGCGGAGAAATTGCTCCAGTACAGGCTTGACGTGGCCTACGTGGCTGCTCAGTTGGCGAAGGTCACAGGACACAAAGGTTATAGGTTTCCATGGGAGAGTGCATACACAGGGGTGGAAGTGACACAGCCATGCTGTCCAGAGGTCGCAGAGTACGAACAGCATATAACCGCCTGCATCTCGTTTGCCGTGAGGCAGTACCTGGCTACCACCAGGAATGAGGAGTGGCTAAAGCACGGCGGATGCGATATAGTTACGAATATAGCTGACTTTTGGGCTTCTAGAGCGGTCATCAATTATAATACTGGACTATACGATATTGAAAGTGAGTACTGTACTTTTTATCATTTTTTATTCCATAGTTGA
Protein
MWRFLTRNKQVAAMMGLLTVAALVVVVMTTKGSEGGHAAANDEEPEDINNDPKVFSTFRLPTNDRFMASIGNGHVAANVFSDTVYMNGLYNGNKGESHRARIPNWANIRLNSTLSHIPYSPVYSLDTKEGAFKVRVDRERSIVTQRIFAHRYYTRAIINQIQVMAKAHAEPNFVRHEIWIAIQLIPGPDSEDIGFKDPVPEIIEGRTVRSTCGQTLKSEDIVYQPLPVEVCVFWTSVPDHLVAPQHGSRVFTFVMTADTNRTVAREEMIRVLQEDGEELYDKHVAEWQKLHQQAHIEIEGNLKVAKVVNAIWYYFLSSLPSEDSHQPLGRFYGLSPTGLARGGTLEDYEGHNFWDTEIWMFPSILLLYPKYAEKLLQYRLDVAYVAAQLAKVTGHKGYRFPWESAYTGVEVTQPCCPEVAEYEQHITACISFAVRQYLATTRNEEWLKHGGCDIVTNIADFWASRAVINYNTGLYDIESEYCTFYHFLFHS
Summary
Uniprot
H9J0L5
A0A2H1VF61
A0A2W1BU94
A0A2A4JJ71
A0A194PTY3
A0A0N0PAQ6
+ More
A0A212FKN6 A0A1B0D4D1 A0A0P6FCS8 A0A0P5MDD1 A0A0P6ALQ4 A0A0P5UAH6 A0A0P4YUX2 Q16LR2 A0A0N8BJJ4 A0A226EFL8 A0A0N8EIH0 A0A1S4FWK3 A0A0P4ZG46 A0A0P5J8L6 A0A0P5JAY0 A0A182MMR2 A0A336L605 A0A1L8E1T7 A0A1J1HZJ3 A0A336LM18 A0A182RWC4 A0A0P5CCP7 A0A182UQT1 A0A3F2Z1W9 A0A084VU32 A0A182Y9R0 A0A182H2Q4 A0A182K1P1 A0A1S4GYL2 A0A1B0A4F2 C3Y7U9 A0A2C9GS60 A0A182U272 A0A1I8NU53 W8BFN0 Q16G35 B0WPY9 A0A0N8E7R4 A0A1A9V831 A0A1A9WXQ2 Q16LR1 A0A182L010 A0A182NT82 B4Q3B6 A0A0J9TLQ3 A0A1D2NEE5 A0A182H2Q5 A0A0K8VNS3 A0A182GBY4 A0A2M4BE19 E9GXQ6 A0A2M4BE20 A0A2M4BEB7 A0A1I8NDG6 Q9VKD9 B3N401 B4P1V6 A0A0P5K136 A0A0P5T5A4 A0A182UFC7 A0A1B0D089 A0A034VJ77 A0A182VKB4 A0A182WRT2 A0A182IXF6 A0A1B0AYY3 A0A2M3Z9W5 A0A2M3Z9H5 B4IE35 A0A1S4HDS5 A0A2C9GS62 A0A182QH94 W5JRD0 W5JSH9 A0A182IBR9 A0A182L007 A0A1S4GYK4 A0A1W4UYB2 A0A2M4AD15 A0A2M4ACX0 A0A1A9XEI8 A0A182L008 A0A182WRT3 A0A182P4M5 W5JSY1 A0A182V5Y9 A0A182K3N7 A0A182U532 A0A182VSG9 A0A1D2NH27 A0A182F1S1 A0NE22 A0A0L0CGD2
A0A212FKN6 A0A1B0D4D1 A0A0P6FCS8 A0A0P5MDD1 A0A0P6ALQ4 A0A0P5UAH6 A0A0P4YUX2 Q16LR2 A0A0N8BJJ4 A0A226EFL8 A0A0N8EIH0 A0A1S4FWK3 A0A0P4ZG46 A0A0P5J8L6 A0A0P5JAY0 A0A182MMR2 A0A336L605 A0A1L8E1T7 A0A1J1HZJ3 A0A336LM18 A0A182RWC4 A0A0P5CCP7 A0A182UQT1 A0A3F2Z1W9 A0A084VU32 A0A182Y9R0 A0A182H2Q4 A0A182K1P1 A0A1S4GYL2 A0A1B0A4F2 C3Y7U9 A0A2C9GS60 A0A182U272 A0A1I8NU53 W8BFN0 Q16G35 B0WPY9 A0A0N8E7R4 A0A1A9V831 A0A1A9WXQ2 Q16LR1 A0A182L010 A0A182NT82 B4Q3B6 A0A0J9TLQ3 A0A1D2NEE5 A0A182H2Q5 A0A0K8VNS3 A0A182GBY4 A0A2M4BE19 E9GXQ6 A0A2M4BE20 A0A2M4BEB7 A0A1I8NDG6 Q9VKD9 B3N401 B4P1V6 A0A0P5K136 A0A0P5T5A4 A0A182UFC7 A0A1B0D089 A0A034VJ77 A0A182VKB4 A0A182WRT2 A0A182IXF6 A0A1B0AYY3 A0A2M3Z9W5 A0A2M3Z9H5 B4IE35 A0A1S4HDS5 A0A2C9GS62 A0A182QH94 W5JRD0 W5JSH9 A0A182IBR9 A0A182L007 A0A1S4GYK4 A0A1W4UYB2 A0A2M4AD15 A0A2M4ACX0 A0A1A9XEI8 A0A182L008 A0A182WRT3 A0A182P4M5 W5JSY1 A0A182V5Y9 A0A182K3N7 A0A182U532 A0A182VSG9 A0A1D2NH27 A0A182F1S1 A0NE22 A0A0L0CGD2
Pubmed
EMBL
BABH01010234
ODYU01002037
SOQ39042.1
KZ150000
PZC75313.1
NWSH01001202
+ More
PCG72145.1 KQ459598 KPI94595.1 KQ461183 KPJ07569.1 AGBW02008010 OWR54304.1 AJVK01011368 GDIQ01051918 GDIQ01051917 JAN42819.1 GDIQ01251042 GDIQ01251041 GDIQ01165561 GDIQ01034765 JAK86164.1 GDIP01027512 JAM76203.1 GDIP01133101 JAL70613.1 GDIP01221877 JAJ01525.1 CH477896 EAT35267.1 GDIQ01171507 JAK80218.1 LNIX01000004 OXA56048.1 GDIQ01023508 JAN71229.1 GDIP01213127 JAJ10275.1 GDIQ01209543 GDIQ01209542 JAK42183.1 GDIQ01208390 JAK43335.1 AXCM01005503 UFQS01001983 UFQT01001983 SSX12854.1 SSX32296.1 GFDF01001417 JAV12667.1 CVRI01000037 CRK93379.1 UFQT01000047 SSX18950.1 GDIP01172424 JAJ50978.1 ATLV01016628 KE525098 KFB41476.1 JXUM01003373 JXUM01003374 JXUM01003375 JXUM01003376 KQ560157 KXJ84127.1 AAAB01008944 GG666490 EEN63604.1 APCN01000730 GAMC01010811 GAMC01010809 GAMC01010807 GAMC01010805 JAB95748.1 CH478339 EAT33196.1 DS232031 EDS32579.1 GDIQ01053557 GDIQ01053556 JAN41181.1 EAT35268.1 CM000361 EDX04747.1 CM002910 KMY89845.1 LJIJ01000067 ODN03634.1 JXUM01003377 KXJ84128.1 GDHF01011811 JAI40503.1 JXUM01159108 KQ573242 KXJ67946.1 GGFJ01002158 MBW51299.1 GL732573 EFX75773.1 GGFJ01002159 MBW51300.1 GGFJ01002160 MBW51301.1 AE014134 BT120276 AAF53137.3 ADC27646.2 CH954177 EDV58853.2 CM000157 EDW88127.2 KRJ97603.1 GDIQ01196296 JAK55429.1 GDIQ01096382 JAL55344.1 AJVK01002273 AJVK01002274 GAKP01017358 GAKP01017354 GAKP01017348 JAC41598.1 JXJN01006030 GGFM01004514 MBW25265.1 GGFM01004401 MBW25152.1 CH480831 EDW45862.1 AXCN02000725 ADMH02000567 ETN65863.1 ETN65865.1 APCN01000729 GGFK01005177 MBW38498.1 GGFK01005298 MBW38619.1 ETN65864.1 LJIJ01000041 ODN04561.1 EAU76693.2 JRES01000438 KNC31305.1
PCG72145.1 KQ459598 KPI94595.1 KQ461183 KPJ07569.1 AGBW02008010 OWR54304.1 AJVK01011368 GDIQ01051918 GDIQ01051917 JAN42819.1 GDIQ01251042 GDIQ01251041 GDIQ01165561 GDIQ01034765 JAK86164.1 GDIP01027512 JAM76203.1 GDIP01133101 JAL70613.1 GDIP01221877 JAJ01525.1 CH477896 EAT35267.1 GDIQ01171507 JAK80218.1 LNIX01000004 OXA56048.1 GDIQ01023508 JAN71229.1 GDIP01213127 JAJ10275.1 GDIQ01209543 GDIQ01209542 JAK42183.1 GDIQ01208390 JAK43335.1 AXCM01005503 UFQS01001983 UFQT01001983 SSX12854.1 SSX32296.1 GFDF01001417 JAV12667.1 CVRI01000037 CRK93379.1 UFQT01000047 SSX18950.1 GDIP01172424 JAJ50978.1 ATLV01016628 KE525098 KFB41476.1 JXUM01003373 JXUM01003374 JXUM01003375 JXUM01003376 KQ560157 KXJ84127.1 AAAB01008944 GG666490 EEN63604.1 APCN01000730 GAMC01010811 GAMC01010809 GAMC01010807 GAMC01010805 JAB95748.1 CH478339 EAT33196.1 DS232031 EDS32579.1 GDIQ01053557 GDIQ01053556 JAN41181.1 EAT35268.1 CM000361 EDX04747.1 CM002910 KMY89845.1 LJIJ01000067 ODN03634.1 JXUM01003377 KXJ84128.1 GDHF01011811 JAI40503.1 JXUM01159108 KQ573242 KXJ67946.1 GGFJ01002158 MBW51299.1 GL732573 EFX75773.1 GGFJ01002159 MBW51300.1 GGFJ01002160 MBW51301.1 AE014134 BT120276 AAF53137.3 ADC27646.2 CH954177 EDV58853.2 CM000157 EDW88127.2 KRJ97603.1 GDIQ01196296 JAK55429.1 GDIQ01096382 JAL55344.1 AJVK01002273 AJVK01002274 GAKP01017358 GAKP01017354 GAKP01017348 JAC41598.1 JXJN01006030 GGFM01004514 MBW25265.1 GGFM01004401 MBW25152.1 CH480831 EDW45862.1 AXCN02000725 ADMH02000567 ETN65863.1 ETN65865.1 APCN01000729 GGFK01005177 MBW38498.1 GGFK01005298 MBW38619.1 ETN65864.1 LJIJ01000041 ODN04561.1 EAU76693.2 JRES01000438 KNC31305.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000092462
+ More
UP000008820 UP000198287 UP000075883 UP000183832 UP000075900 UP000075903 UP000076407 UP000030765 UP000076408 UP000069940 UP000249989 UP000075881 UP000092445 UP000001554 UP000075840 UP000075902 UP000095300 UP000002320 UP000078200 UP000091820 UP000075882 UP000075884 UP000000304 UP000094527 UP000000305 UP000095301 UP000000803 UP000008711 UP000002282 UP000075880 UP000092460 UP000001292 UP000075886 UP000000673 UP000192221 UP000092443 UP000075885 UP000075920 UP000069272 UP000007062 UP000037069
UP000008820 UP000198287 UP000075883 UP000183832 UP000075900 UP000075903 UP000076407 UP000030765 UP000076408 UP000069940 UP000249989 UP000075881 UP000092445 UP000001554 UP000075840 UP000075902 UP000095300 UP000002320 UP000078200 UP000091820 UP000075882 UP000075884 UP000000304 UP000094527 UP000000305 UP000095301 UP000000803 UP000008711 UP000002282 UP000075880 UP000092460 UP000001292 UP000075886 UP000000673 UP000192221 UP000092443 UP000075885 UP000075920 UP000069272 UP000007062 UP000037069
Interpro
Gene 3D
ProteinModelPortal
H9J0L5
A0A2H1VF61
A0A2W1BU94
A0A2A4JJ71
A0A194PTY3
A0A0N0PAQ6
+ More
A0A212FKN6 A0A1B0D4D1 A0A0P6FCS8 A0A0P5MDD1 A0A0P6ALQ4 A0A0P5UAH6 A0A0P4YUX2 Q16LR2 A0A0N8BJJ4 A0A226EFL8 A0A0N8EIH0 A0A1S4FWK3 A0A0P4ZG46 A0A0P5J8L6 A0A0P5JAY0 A0A182MMR2 A0A336L605 A0A1L8E1T7 A0A1J1HZJ3 A0A336LM18 A0A182RWC4 A0A0P5CCP7 A0A182UQT1 A0A3F2Z1W9 A0A084VU32 A0A182Y9R0 A0A182H2Q4 A0A182K1P1 A0A1S4GYL2 A0A1B0A4F2 C3Y7U9 A0A2C9GS60 A0A182U272 A0A1I8NU53 W8BFN0 Q16G35 B0WPY9 A0A0N8E7R4 A0A1A9V831 A0A1A9WXQ2 Q16LR1 A0A182L010 A0A182NT82 B4Q3B6 A0A0J9TLQ3 A0A1D2NEE5 A0A182H2Q5 A0A0K8VNS3 A0A182GBY4 A0A2M4BE19 E9GXQ6 A0A2M4BE20 A0A2M4BEB7 A0A1I8NDG6 Q9VKD9 B3N401 B4P1V6 A0A0P5K136 A0A0P5T5A4 A0A182UFC7 A0A1B0D089 A0A034VJ77 A0A182VKB4 A0A182WRT2 A0A182IXF6 A0A1B0AYY3 A0A2M3Z9W5 A0A2M3Z9H5 B4IE35 A0A1S4HDS5 A0A2C9GS62 A0A182QH94 W5JRD0 W5JSH9 A0A182IBR9 A0A182L007 A0A1S4GYK4 A0A1W4UYB2 A0A2M4AD15 A0A2M4ACX0 A0A1A9XEI8 A0A182L008 A0A182WRT3 A0A182P4M5 W5JSY1 A0A182V5Y9 A0A182K3N7 A0A182U532 A0A182VSG9 A0A1D2NH27 A0A182F1S1 A0NE22 A0A0L0CGD2
A0A212FKN6 A0A1B0D4D1 A0A0P6FCS8 A0A0P5MDD1 A0A0P6ALQ4 A0A0P5UAH6 A0A0P4YUX2 Q16LR2 A0A0N8BJJ4 A0A226EFL8 A0A0N8EIH0 A0A1S4FWK3 A0A0P4ZG46 A0A0P5J8L6 A0A0P5JAY0 A0A182MMR2 A0A336L605 A0A1L8E1T7 A0A1J1HZJ3 A0A336LM18 A0A182RWC4 A0A0P5CCP7 A0A182UQT1 A0A3F2Z1W9 A0A084VU32 A0A182Y9R0 A0A182H2Q4 A0A182K1P1 A0A1S4GYL2 A0A1B0A4F2 C3Y7U9 A0A2C9GS60 A0A182U272 A0A1I8NU53 W8BFN0 Q16G35 B0WPY9 A0A0N8E7R4 A0A1A9V831 A0A1A9WXQ2 Q16LR1 A0A182L010 A0A182NT82 B4Q3B6 A0A0J9TLQ3 A0A1D2NEE5 A0A182H2Q5 A0A0K8VNS3 A0A182GBY4 A0A2M4BE19 E9GXQ6 A0A2M4BE20 A0A2M4BEB7 A0A1I8NDG6 Q9VKD9 B3N401 B4P1V6 A0A0P5K136 A0A0P5T5A4 A0A182UFC7 A0A1B0D089 A0A034VJ77 A0A182VKB4 A0A182WRT2 A0A182IXF6 A0A1B0AYY3 A0A2M3Z9W5 A0A2M3Z9H5 B4IE35 A0A1S4HDS5 A0A2C9GS62 A0A182QH94 W5JRD0 W5JSH9 A0A182IBR9 A0A182L007 A0A1S4GYK4 A0A1W4UYB2 A0A2M4AD15 A0A2M4ACX0 A0A1A9XEI8 A0A182L008 A0A182WRT3 A0A182P4M5 W5JSY1 A0A182V5Y9 A0A182K3N7 A0A182U532 A0A182VSG9 A0A1D2NH27 A0A182F1S1 A0NE22 A0A0L0CGD2
PDB
3WIR
E-value=1.4654e-25,
Score=289
Ontologies
GO
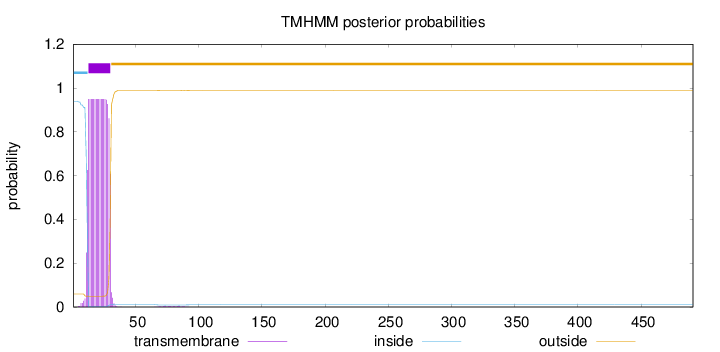
Topology
Length:
491
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.8499099999999
Exp number, first 60 AAs:
17.75741
Total prob of N-in:
0.94022
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 30
outside
31 - 491
Population Genetic Test Statistics
Pi
300.07103
Theta
202.4607
Tajima's D
0.701059
CLR
0.780589
CSRT
0.570121493925304
Interpretation
Uncertain
