Gene
KWMTBOMO02248
Pre Gene Modal
BGIBMGA003104
Annotation
Chymotrypsin-1_[Operophtera_brumata]
Full name
Chymotrypsin
+ More
Trypsin
Trypsin
Location in the cell
Mitochondrial Reliability : 1.194
Sequence
CDS
ATGTCTGGTTTTATTGTGGGCGGATATTTGGCGTCAATCAAGGAATATCCCCACTCAGCTTTTTTGAGAGCCGGTTGCGTCACACCAACCGGTAATGTCCGCAAATGGCAATGTGGATCATCGATAATCAATCAGATGATGTTGCTGACCGCGGCTCATTGCTTAGAAGAGGCAAAGGAAATTCTCGTTACTGTCGGAAGTGAAAACAAAAATATGGGTTTGACAATAAAAGCTGGAAGATACATTATACATGAAAATTATGATCCGTCGGCGTTACATTCGGATCTAGGTCTCATTGGGCTATCTAAGCCGTTGAAATTCACTTCGAAAATACAACGAGTAGCCCTGATGAGGAGCCCGCCTTATAAAGAAAAAGCATTTGTCGCTGGTTGGGGAGTGGTTGACGAAATCAATATCAGAGTGATTCCTGTGTTAAAATATTTGGATCAAATTGTCCGAAATCAGAATAGCTGCTACAAAGAAAAAATCTATGAAGCAATACCAGGAACCATTTGCGCTAGCAATGCAAAAGCCTCAGAATATCCATCTATGTAA
Protein
MSGFIVGGYLASIKEYPHSAFLRAGCVTPTGNVRKWQCGSSIINQMMLLTAAHCLEEAKEILVTVGSENKNMGLTIKAGRYIIHENYDPSALHSDLGLIGLSKPLKFTSKIQRVALMRSPPYKEKAFVAGWGVVDEINIRVIPVLKYLDQIVRNQNSCYKEKIYEAIPGTICASNAKASEYPSM
Summary
Description
Serine protease with chymotryptic and collagenolytic activities.
Catalytic Activity
Preferential cleavage: Tyr-|-Xaa, Trp-|-Xaa, Phe-|-Xaa, Leu-|-Xaa.
Preferential cleavage: Arg-|-Xaa, Lys-|-Xaa.
Preferential cleavage: Arg-|-Xaa, Lys-|-Xaa.
Similarity
Belongs to the peptidase S1 family.
Keywords
Collagen degradation
Digestion
Disulfide bond
Glycoprotein
Hydrolase
Protease
Secreted
Serine protease
Signal
Zymogen
Direct protein sequencing
Feature
propeptide Activation peptide
chain Chymotrypsin
chain Chymotrypsin
Uniprot
H9J0R7
A0A2H1WKW7
A0A2H1VR93
A0A2W1BJQ7
A0A0L7LFE7
A0A0L7KT34
+ More
A0A2A4JL27 A0A0L7LF03 A0A212FKP7 A0A194RKI2 H9J0R6 A0A2H1VHC8 A0A194QDC4 A0A0L7LG02 A0A0L7KVC9 A0A0L7LF91 A0A2H1V9I9 A0A194Q913 A0A212FKM8 A0A2H1W4Q5 A0A1A9WKQ0 A0A0L0BSQ2 A0A1B0B1H8 D6WPA6 A0A1A9XWP6 A0A336MUY7 A0A336MXC4 A0A182PK66 T1H2V5 O97398 A0A2D9XLI4 B4JVV3 E0W3U6 A0A1I8M656 A0A1A9Z6S9 Q6XHL5 A1XG66 B5APZ4 B5AQM2 A0A2P2I159 A0A0L0C9D3 A0A182QJV9 B4PJL5 A1XG65 A0NG43 B4J3I4 A0A182KJU0 A0A182IGM8 F1DQG5 D6WZJ8 A0A182IN32 B4KZJ9 O15944 A0A2A3E9B5 A0A182UJ75 A0A2P2I1H3 E1ZZV4 T1GX22 T1GQB9 Q8T4A8 B4HKU3 A0A0A1WN98 D3TMZ8 A0A3G5BIF4 Q28Y22 A7UNZ4 A0A1I8M4V1 A0A084W5I8 A0A1I8M4V2 Q16ZF3 Q25081 T1P9U9 A0A182RAB0 B4JBA6 E9GE64 A7DYX5 A0A3G5BIH9 B4GGG2 P51588 A0A1B0FLW7 B3M4K2 B3NHT6 V5G1I3 D6WN49 A0A1I8M2D3 Q8I8E2 A0A1B3TNZ7 A0A2E4J7M2 A0A182MAA4 A0A0K8WL22 A0A182H9P0 A0A182K9L9 A0A099ZWT2 Q52NW5 A0A1I8NW71 A0A1I8QDI6 A0A182Q608 A0A232ET09 Q4V440
A0A2A4JL27 A0A0L7LF03 A0A212FKP7 A0A194RKI2 H9J0R6 A0A2H1VHC8 A0A194QDC4 A0A0L7LG02 A0A0L7KVC9 A0A0L7LF91 A0A2H1V9I9 A0A194Q913 A0A212FKM8 A0A2H1W4Q5 A0A1A9WKQ0 A0A0L0BSQ2 A0A1B0B1H8 D6WPA6 A0A1A9XWP6 A0A336MUY7 A0A336MXC4 A0A182PK66 T1H2V5 O97398 A0A2D9XLI4 B4JVV3 E0W3U6 A0A1I8M656 A0A1A9Z6S9 Q6XHL5 A1XG66 B5APZ4 B5AQM2 A0A2P2I159 A0A0L0C9D3 A0A182QJV9 B4PJL5 A1XG65 A0NG43 B4J3I4 A0A182KJU0 A0A182IGM8 F1DQG5 D6WZJ8 A0A182IN32 B4KZJ9 O15944 A0A2A3E9B5 A0A182UJ75 A0A2P2I1H3 E1ZZV4 T1GX22 T1GQB9 Q8T4A8 B4HKU3 A0A0A1WN98 D3TMZ8 A0A3G5BIF4 Q28Y22 A7UNZ4 A0A1I8M4V1 A0A084W5I8 A0A1I8M4V2 Q16ZF3 Q25081 T1P9U9 A0A182RAB0 B4JBA6 E9GE64 A7DYX5 A0A3G5BIH9 B4GGG2 P51588 A0A1B0FLW7 B3M4K2 B3NHT6 V5G1I3 D6WN49 A0A1I8M2D3 Q8I8E2 A0A1B3TNZ7 A0A2E4J7M2 A0A182MAA4 A0A0K8WL22 A0A182H9P0 A0A182K9L9 A0A099ZWT2 Q52NW5 A0A1I8NW71 A0A1I8QDI6 A0A182Q608 A0A232ET09 Q4V440
EC Number
3.4.21.1
3.4.21.4
3.4.21.4
Pubmed
19121390
28756777
26227816
22118469
26354079
26108605
+ More
18362917 19820115 10612046 29337314 17994087 20566863 25315136 14525923 17651235 17550304 12364791 20966253 22509400 9295327 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 20353571 30400621 15632085 24438588 17510324 7808473 21292972 17573377 8620885 27553646 26483478 17502004 28648823
18362917 19820115 10612046 29337314 17994087 20566863 25315136 14525923 17651235 17550304 12364791 20966253 22509400 9295327 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 20353571 30400621 15632085 24438588 17510324 7808473 21292972 17573377 8620885 27553646 26483478 17502004 28648823
EMBL
BABH01010245
ODYU01009360
SOQ53730.1
ODYU01003944
SOQ43317.1
KZ150000
+ More
PZC75312.1 JTDY01001347 KOB74130.1 JTDY01006244 KOB66174.1 NWSH01001202 PCG72143.1 KOB74133.1 AGBW02008010 OWR54306.1 KQ460045 KPJ18047.1 BABH01010242 BABH01010243 ODYU01002383 SOQ39812.1 KQ459324 KPJ01456.1 KOB74131.1 JTDY01005274 KOB67168.1 KOB74132.1 ODYU01001393 SOQ37513.1 KPJ01455.1 OWR54305.1 ODYU01006324 SOQ48069.1 JRES01001422 KNC23046.1 JXJN01007177 KQ971354 EFA07583.1 UFQT01002465 SSX33525.1 UFQS01003363 UFQT01003363 SSX15542.1 SSX34906.1 CAQQ02171618 CAQQ02171619 Y17904 CAA76928.1 NZWM01000032 MAQ69946.1 CH916375 EDV98091.1 DS235883 EEB20302.1 AY232168 AAR10191.1 DQ356025 ABC88740.1 EU869219 ACF93734.1 EU869872 ACF98290.1 IACF01002059 LAB67729.1 JRES01000827 KNC28039.1 AXCN02001863 CM000159 EDW94703.1 DQ356024 ABC88739.1 AAAB01008980 EAU76138.2 CH916366 EDV96186.1 APCN01005255 HQ878386 ADY17978.1 KQ971372 EFA10697.1 CH933809 EDW17926.1 AB002407 BAA22400.1 KZ288316 PBC28310.1 IACF01002060 LAB67730.1 GL435530 EFN73265.1 CAQQ02388900 CAQQ02035930 AE014296 AY089278 AAF49326.2 AAL90016.1 CH480815 EDW41898.1 GBXI01013758 JAD00534.1 EZ422800 ADD19076.1 MK075162 AYV99565.1 CM000071 EAL26143.1 EU095344 ABU49589.1 ATLV01020656 KE525304 KFB45482.1 CH477491 EAT40037.1 X74304 CAA52357.1 KA645497 AFP60126.1 CH916368 EDW02911.1 GL732540 EFX82212.1 AM765877 CAO78778.1 MK075184 AYV99587.1 CH479183 EDW35582.1 X94691 CCAG010022336 CCAG010022337 CCAG010022338 CH902618 EDV40496.2 CH954178 EDV51951.1 GALX01004586 JAB63880.1 KQ971342 EFA04551.1 AY155564 AAN75002.1 KU925752 AOG75377.1 PAVX01000020 MBE37760.1 AXCM01001005 GDHF01000734 JAI51580.1 JXUM01030427 KQ560884 KXJ80485.1 KL869824 KGL86182.1 AY995188 AAX94742.1 AXCN02000888 NNAY01002348 OXU21493.1 BT023166 AAY55582.1
PZC75312.1 JTDY01001347 KOB74130.1 JTDY01006244 KOB66174.1 NWSH01001202 PCG72143.1 KOB74133.1 AGBW02008010 OWR54306.1 KQ460045 KPJ18047.1 BABH01010242 BABH01010243 ODYU01002383 SOQ39812.1 KQ459324 KPJ01456.1 KOB74131.1 JTDY01005274 KOB67168.1 KOB74132.1 ODYU01001393 SOQ37513.1 KPJ01455.1 OWR54305.1 ODYU01006324 SOQ48069.1 JRES01001422 KNC23046.1 JXJN01007177 KQ971354 EFA07583.1 UFQT01002465 SSX33525.1 UFQS01003363 UFQT01003363 SSX15542.1 SSX34906.1 CAQQ02171618 CAQQ02171619 Y17904 CAA76928.1 NZWM01000032 MAQ69946.1 CH916375 EDV98091.1 DS235883 EEB20302.1 AY232168 AAR10191.1 DQ356025 ABC88740.1 EU869219 ACF93734.1 EU869872 ACF98290.1 IACF01002059 LAB67729.1 JRES01000827 KNC28039.1 AXCN02001863 CM000159 EDW94703.1 DQ356024 ABC88739.1 AAAB01008980 EAU76138.2 CH916366 EDV96186.1 APCN01005255 HQ878386 ADY17978.1 KQ971372 EFA10697.1 CH933809 EDW17926.1 AB002407 BAA22400.1 KZ288316 PBC28310.1 IACF01002060 LAB67730.1 GL435530 EFN73265.1 CAQQ02388900 CAQQ02035930 AE014296 AY089278 AAF49326.2 AAL90016.1 CH480815 EDW41898.1 GBXI01013758 JAD00534.1 EZ422800 ADD19076.1 MK075162 AYV99565.1 CM000071 EAL26143.1 EU095344 ABU49589.1 ATLV01020656 KE525304 KFB45482.1 CH477491 EAT40037.1 X74304 CAA52357.1 KA645497 AFP60126.1 CH916368 EDW02911.1 GL732540 EFX82212.1 AM765877 CAO78778.1 MK075184 AYV99587.1 CH479183 EDW35582.1 X94691 CCAG010022336 CCAG010022337 CCAG010022338 CH902618 EDV40496.2 CH954178 EDV51951.1 GALX01004586 JAB63880.1 KQ971342 EFA04551.1 AY155564 AAN75002.1 KU925752 AOG75377.1 PAVX01000020 MBE37760.1 AXCM01001005 GDHF01000734 JAI51580.1 JXUM01030427 KQ560884 KXJ80485.1 KL869824 KGL86182.1 AY995188 AAX94742.1 AXCN02000888 NNAY01002348 OXU21493.1 BT023166 AAY55582.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000091820 UP000037069 UP000092460 UP000007266 UP000092443 UP000075885 UP000015102 UP000001070 UP000009046 UP000095301 UP000092445 UP000075886 UP000002282 UP000007062 UP000075882 UP000075840 UP000075880 UP000009192 UP000242457 UP000075902 UP000000311 UP000000803 UP000001292 UP000001819 UP000030765 UP000008820 UP000075900 UP000000305 UP000008744 UP000092444 UP000007801 UP000008711 UP000075883 UP000069940 UP000249989 UP000075881 UP000053858 UP000095300 UP000215335
UP000091820 UP000037069 UP000092460 UP000007266 UP000092443 UP000075885 UP000015102 UP000001070 UP000009046 UP000095301 UP000092445 UP000075886 UP000002282 UP000007062 UP000075882 UP000075840 UP000075880 UP000009192 UP000242457 UP000075902 UP000000311 UP000000803 UP000001292 UP000001819 UP000030765 UP000008820 UP000075900 UP000000305 UP000008744 UP000092444 UP000007801 UP000008711 UP000075883 UP000069940 UP000249989 UP000075881 UP000053858 UP000095300 UP000215335
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J0R7
A0A2H1WKW7
A0A2H1VR93
A0A2W1BJQ7
A0A0L7LFE7
A0A0L7KT34
+ More
A0A2A4JL27 A0A0L7LF03 A0A212FKP7 A0A194RKI2 H9J0R6 A0A2H1VHC8 A0A194QDC4 A0A0L7LG02 A0A0L7KVC9 A0A0L7LF91 A0A2H1V9I9 A0A194Q913 A0A212FKM8 A0A2H1W4Q5 A0A1A9WKQ0 A0A0L0BSQ2 A0A1B0B1H8 D6WPA6 A0A1A9XWP6 A0A336MUY7 A0A336MXC4 A0A182PK66 T1H2V5 O97398 A0A2D9XLI4 B4JVV3 E0W3U6 A0A1I8M656 A0A1A9Z6S9 Q6XHL5 A1XG66 B5APZ4 B5AQM2 A0A2P2I159 A0A0L0C9D3 A0A182QJV9 B4PJL5 A1XG65 A0NG43 B4J3I4 A0A182KJU0 A0A182IGM8 F1DQG5 D6WZJ8 A0A182IN32 B4KZJ9 O15944 A0A2A3E9B5 A0A182UJ75 A0A2P2I1H3 E1ZZV4 T1GX22 T1GQB9 Q8T4A8 B4HKU3 A0A0A1WN98 D3TMZ8 A0A3G5BIF4 Q28Y22 A7UNZ4 A0A1I8M4V1 A0A084W5I8 A0A1I8M4V2 Q16ZF3 Q25081 T1P9U9 A0A182RAB0 B4JBA6 E9GE64 A7DYX5 A0A3G5BIH9 B4GGG2 P51588 A0A1B0FLW7 B3M4K2 B3NHT6 V5G1I3 D6WN49 A0A1I8M2D3 Q8I8E2 A0A1B3TNZ7 A0A2E4J7M2 A0A182MAA4 A0A0K8WL22 A0A182H9P0 A0A182K9L9 A0A099ZWT2 Q52NW5 A0A1I8NW71 A0A1I8QDI6 A0A182Q608 A0A232ET09 Q4V440
A0A2A4JL27 A0A0L7LF03 A0A212FKP7 A0A194RKI2 H9J0R6 A0A2H1VHC8 A0A194QDC4 A0A0L7LG02 A0A0L7KVC9 A0A0L7LF91 A0A2H1V9I9 A0A194Q913 A0A212FKM8 A0A2H1W4Q5 A0A1A9WKQ0 A0A0L0BSQ2 A0A1B0B1H8 D6WPA6 A0A1A9XWP6 A0A336MUY7 A0A336MXC4 A0A182PK66 T1H2V5 O97398 A0A2D9XLI4 B4JVV3 E0W3U6 A0A1I8M656 A0A1A9Z6S9 Q6XHL5 A1XG66 B5APZ4 B5AQM2 A0A2P2I159 A0A0L0C9D3 A0A182QJV9 B4PJL5 A1XG65 A0NG43 B4J3I4 A0A182KJU0 A0A182IGM8 F1DQG5 D6WZJ8 A0A182IN32 B4KZJ9 O15944 A0A2A3E9B5 A0A182UJ75 A0A2P2I1H3 E1ZZV4 T1GX22 T1GQB9 Q8T4A8 B4HKU3 A0A0A1WN98 D3TMZ8 A0A3G5BIF4 Q28Y22 A7UNZ4 A0A1I8M4V1 A0A084W5I8 A0A1I8M4V2 Q16ZF3 Q25081 T1P9U9 A0A182RAB0 B4JBA6 E9GE64 A7DYX5 A0A3G5BIH9 B4GGG2 P51588 A0A1B0FLW7 B3M4K2 B3NHT6 V5G1I3 D6WN49 A0A1I8M2D3 Q8I8E2 A0A1B3TNZ7 A0A2E4J7M2 A0A182MAA4 A0A0K8WL22 A0A182H9P0 A0A182K9L9 A0A099ZWT2 Q52NW5 A0A1I8NW71 A0A1I8QDI6 A0A182Q608 A0A232ET09 Q4V440
PDB
3TGJ
E-value=1.55785e-11,
Score=163
Ontologies
GO
Topology
Subcellular location
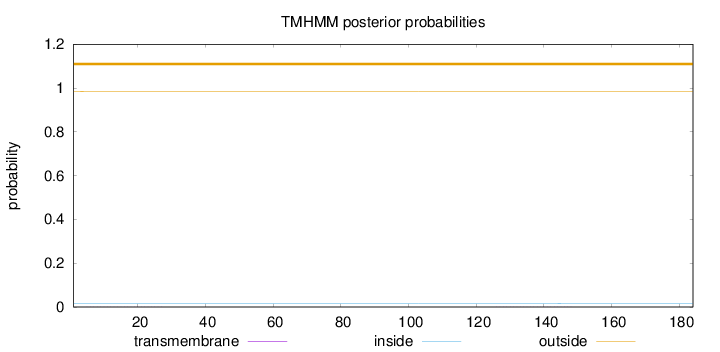
Length:
184
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0154
Exp number, first 60 AAs:
0.00531999999999999
Total prob of N-in:
0.01545
outside
1 - 184
Population Genetic Test Statistics
Pi
120.7047
Theta
109.458241
Tajima's D
0.322364
CLR
60.746685
CSRT
0.464776761161942
Interpretation
Uncertain
