Gene
KWMTBOMO02247
Pre Gene Modal
BGIBMGA003105
Annotation
PREDICTED:_trypsin_beta-like_[Bombyx_mori]
Full name
Inactive serine protease scarface
Location in the cell
Extracellular Reliability : 1.587 PlasmaMembrane Reliability : 1.483
Sequence
CDS
ATGAATAATCTGTTGTTATATTTATACGTTTTAACTGTCCTCGGGCCATTTGCTTTATCGAAACACACAAATGTTACTAAAGATTTGGAACCTCTGATTTACAAAGGATACGAAGTTGGAATACACGATTATTCCTACGCTTGTTTTCTGCATTTCAAAACCACCAGGCCGACTTCGTGTGGAGCTTCCCTTATAATGGTCCAAGCTGTTCTGACGGCAGCGCACTGCATGGACGATATCCGGATCAAGAGCGGCGAAATTCACGCGTTTTTCGGATCAAACGTACCAAAAGATTCTTCGGTTATCAGAAGAGTCGGGAATTATGAAATAAATCCAAAATATAATGAGGTGAATGGTAAGAACAATCTGGCAGTGGCGTTTCTGGATTATAGAGTACCACTTCGGCAGGGCATCAAGAAGATCCCGCTGTCAATCAGAGATCCAGTGCCACGAACTGATCTGTTTTCAGTGGGTTGGGGCAAACAAAGCGTAAGTAAACTATATTCTCAATTTCTACATTAG
Protein
MNNLLLYLYVLTVLGPFALSKHTNVTKDLEPLIYKGYEVGIHDYSYACFLHFKTTRPTSCGASLIMVQAVLTAAHCMDDIRIKSGEIHAFFGSNVPKDSSVIRRVGNYEINPKYNEVNGKNNLAVAFLDYRVPLRQGIKKIPLSIRDPVPRTDLFSVGWGKQSVSKLYSQFLH
Summary
Description
Inactive serine protease that plays a role in germ-band retraction and dorsal closure morphogenesis in embryogenesis; contributes to amnioserosa attachment and epithelial apico-basal polarity by regulating the localization of laminin LanA on the apical side of the amnioserosa epithelium (PubMed:28628612, PubMed:20379222). Contributes to epithelial morphogenesis probably by regulating the bsk/JNK pathway, as part of a negative-feedback loop, and by modulating the cross-talk between the Egfr, bsk/JNK and dpp signal transduction pathways (PubMed:28628612, PubMed:20379222). In larval development, antagonizes the morphogenetic movements controlled by the bsk/JNK signaling including male genitalia formation and thorax development (PubMed:20379222, PubMed:20530545, PubMed:25737837).
Similarity
Belongs to the peptidase S1 family.
Keywords
Complete proteome
Developmental protein
Disulfide bond
Reference proteome
Secreted
Serine protease homolog
Signal
Feature
chain Inactive serine protease scarface
Uniprot
D9HQ67
D9HQ25
A0A212FKN2
T1GDM2
R4G2C0
R4G7C9
+ More
T1GQB9 B4KXH6 A0A0U5GTC1 A0A1I8MTT1 T1P882 A0A1B0BUI5 G4YJG1 B6Z1X6 A0A1Q1NPF7 W5LWA6 A0A1W4V7B4 A0A1Q1NPF4 A0A1Q1NPI1 A0A1Q1NP77 A0A1W4V5K3 A0A0K8SCH7 A0A146LZD5 A0A034VGE4 A0A0A9W4V0 A0A1A9WYS8 B4N135 A0A034VKN4 A0A3P8NRS8 A0A3P8NRG2 A0A1A9XIV4 A0A1B0FAF0 A0A2V1DTG0 A0A1I8NNQ7 A0A1A9US87 A0A182FUP8 A0A1A9ZAW2 R0JV69 B3MBQ9 A0A218UA67 B4L9R3 T1GQ14 A0A0P9C129 W5LXF7 A7DYW0 A0A310SCG7 Q0GSS5 A7DYV6 Q0GSS6 Q0GSS7 U3IJ08 A0A3P9CA61 A0A0A1XLZ0 A0A2K8JSL6 A0A0K8WBX2 I3K9X1 A0A1D5P7Y3 A7DYV9 A7DYW5 K7FZ69 Q0GST3 Q0GST5 A0A0J9R7H1 A7DYV7 A0A182FUP6 Q7K5M0 Q9VQA0 A0A0J9TTW9 L0ATN6 B4II01 A0A1B6MUX8 I3K8H2 V8NPI1 A0A336LFU1 A0A0L8HT00 A0A3M7R237 A0A182N2U4 A0A1J1I662 A7DYW3 A0A1A7XCF2 A0A3B4AJ83 A0A1A9WAU6 A0A0L7R7Y2
T1GQB9 B4KXH6 A0A0U5GTC1 A0A1I8MTT1 T1P882 A0A1B0BUI5 G4YJG1 B6Z1X6 A0A1Q1NPF7 W5LWA6 A0A1W4V7B4 A0A1Q1NPF4 A0A1Q1NPI1 A0A1Q1NP77 A0A1W4V5K3 A0A0K8SCH7 A0A146LZD5 A0A034VGE4 A0A0A9W4V0 A0A1A9WYS8 B4N135 A0A034VKN4 A0A3P8NRS8 A0A3P8NRG2 A0A1A9XIV4 A0A1B0FAF0 A0A2V1DTG0 A0A1I8NNQ7 A0A1A9US87 A0A182FUP8 A0A1A9ZAW2 R0JV69 B3MBQ9 A0A218UA67 B4L9R3 T1GQ14 A0A0P9C129 W5LXF7 A7DYW0 A0A310SCG7 Q0GSS5 A7DYV6 Q0GSS6 Q0GSS7 U3IJ08 A0A3P9CA61 A0A0A1XLZ0 A0A2K8JSL6 A0A0K8WBX2 I3K9X1 A0A1D5P7Y3 A7DYV9 A7DYW5 K7FZ69 Q0GST3 Q0GST5 A0A0J9R7H1 A7DYV7 A0A182FUP6 Q7K5M0 Q9VQA0 A0A0J9TTW9 L0ATN6 B4II01 A0A1B6MUX8 I3K8H2 V8NPI1 A0A336LFU1 A0A0L8HT00 A0A3M7R237 A0A182N2U4 A0A1J1I662 A7DYW3 A0A1A7XCF2 A0A3B4AJ83 A0A1A9WAU6 A0A0L7R7Y2
Pubmed
20375075
22118469
17994087
25315136
16946064
19294452
+ More
28130397 26823975 25348373 25401762 29679020 17573377 16783023 23749191 25186727 25830018 15592404 17381049 22936249 10731132 12537572 12537569 15342518 20379222 20530545 25737837 28628612 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24297900 30375419 25463417
28130397 26823975 25348373 25401762 29679020 17573377 16783023 23749191 25186727 25830018 15592404 17381049 22936249 10731132 12537572 12537569 15342518 20379222 20530545 25737837 28628612 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24297900 30375419 25463417
EMBL
HM023822
ADJ58555.1
HM023780
ADJ58513.1
AGBW02008010
OWR54307.1
+ More
CAQQ02093592 GAGI01000010 JAA74559.1 GAGI01000011 JAA74558.1 CAQQ02035930 CH933809 EDW18662.2 CDMC01000006 CEN62071.1 KA644802 AFP59431.1 JXJN01020710 JH159151 EGZ29916.1 EU790599 ACJ22645.1 KX459638 AQM58389.1 KX459645 AQM58396.1 KX459663 AQM58414.1 KX459571 AQM58322.1 GBRD01015002 JAG50824.1 GDHC01005675 JAQ12954.1 GAKP01016576 JAC42376.1 GBHO01041168 JAG02436.1 CH963920 EDW78197.1 GAKP01016577 JAC42375.1 CCAG010019386 CCAG010019387 CCAG010019388 KZ805371 PVI00574.1 KB743121 EOB01127.1 CH902619 EDV38205.2 MUZQ01000559 OWK50390.1 CH933816 EDW17111.1 CAQQ02143884 KPU77319.1 AM765862 CAO78763.1 KQ764252 OAD54568.1 DQ539273 DQ539274 DQ539275 DQ539276 DQ539277 DQ539280 AM765864 ABG02794.1 ABG02795.1 ABG02796.1 ABG02797.1 ABG02798.1 ABG02801.1 CAO78765.1 AM765858 CAO78759.1 DQ539279 ABG02800.1 DQ539278 AM765863 ABG02799.1 CAO78764.1 ADON01075126 ADON01075127 ADON01075128 ADON01075129 ADON01075130 ADON01075131 GBXI01001973 JAD12319.1 KY031181 ATU82932.1 GDHF01003626 JAI48688.1 AERX01012107 AERX01012108 AADN05000689 AM765861 CAO78762.1 AM765867 CAO78768.1 AGCU01052657 DQ539271 DQ539272 AM765866 ABG02792.1 ABG02793.1 CAO78767.1 DQ539244 DQ539245 DQ539246 DQ539247 DQ539248 DQ539249 DQ539250 DQ539251 DQ539252 DQ539253 DQ539254 DQ539255 DQ539256 DQ539258 DQ539261 DQ539262 DQ539263 DQ539264 DQ539265 DQ539266 DQ539267 DQ539268 DQ539269 DQ539270 AM765857 AM765868 AM765869 AM765870 AM765871 AM765872 AM765873 AM765874 AM765875 AM765876 ABG02765.1 ABG02766.1 ABG02767.1 ABG02768.1 ABG02769.1 ABG02770.1 ABG02771.1 ABG02772.1 ABG02773.1 ABG02774.1 ABG02775.1 ABG02776.1 ABG02777.1 ABG02779.1 ABG02782.1 ABG02783.1 ABG02784.1 ABG02785.1 ABG02786.1 ABG02787.1 ABG02788.1 ABG02789.1 ABG02790.1 ABG02791.1 CAO78758.1 CAO78769.1 CAO78770.1 CAO78771.1 CAO78772.1 CAO78773.1 CAO78774.1 CAO78775.1 CAO78776.1 CAO78777.1 CM002911 KMY91639.1 AM765859 CAO78760.1 AE013599 AY047548 AAK77280.1 AE014134 AY060796 AAF51277.1 AAL28344.1 KMY91640.1 JX915894 AFZ78860.1 CH480841 EDW49527.1 GEBQ01011698 GEBQ01000257 JAT28279.1 JAT39720.1 AERX01017497 AZIM01002473 ETE63965.1 UFQS01005297 UFQT01005297 SSX16784.1 SSX35971.1 KQ417459 KOF91865.1 REGN01004409 RNA17643.1 CVRI01000038 CRK93881.1 AM765865 CAO78766.1 HADW01014130 SBP15530.1 KQ414637 KOC66990.1
CAQQ02093592 GAGI01000010 JAA74559.1 GAGI01000011 JAA74558.1 CAQQ02035930 CH933809 EDW18662.2 CDMC01000006 CEN62071.1 KA644802 AFP59431.1 JXJN01020710 JH159151 EGZ29916.1 EU790599 ACJ22645.1 KX459638 AQM58389.1 KX459645 AQM58396.1 KX459663 AQM58414.1 KX459571 AQM58322.1 GBRD01015002 JAG50824.1 GDHC01005675 JAQ12954.1 GAKP01016576 JAC42376.1 GBHO01041168 JAG02436.1 CH963920 EDW78197.1 GAKP01016577 JAC42375.1 CCAG010019386 CCAG010019387 CCAG010019388 KZ805371 PVI00574.1 KB743121 EOB01127.1 CH902619 EDV38205.2 MUZQ01000559 OWK50390.1 CH933816 EDW17111.1 CAQQ02143884 KPU77319.1 AM765862 CAO78763.1 KQ764252 OAD54568.1 DQ539273 DQ539274 DQ539275 DQ539276 DQ539277 DQ539280 AM765864 ABG02794.1 ABG02795.1 ABG02796.1 ABG02797.1 ABG02798.1 ABG02801.1 CAO78765.1 AM765858 CAO78759.1 DQ539279 ABG02800.1 DQ539278 AM765863 ABG02799.1 CAO78764.1 ADON01075126 ADON01075127 ADON01075128 ADON01075129 ADON01075130 ADON01075131 GBXI01001973 JAD12319.1 KY031181 ATU82932.1 GDHF01003626 JAI48688.1 AERX01012107 AERX01012108 AADN05000689 AM765861 CAO78762.1 AM765867 CAO78768.1 AGCU01052657 DQ539271 DQ539272 AM765866 ABG02792.1 ABG02793.1 CAO78767.1 DQ539244 DQ539245 DQ539246 DQ539247 DQ539248 DQ539249 DQ539250 DQ539251 DQ539252 DQ539253 DQ539254 DQ539255 DQ539256 DQ539258 DQ539261 DQ539262 DQ539263 DQ539264 DQ539265 DQ539266 DQ539267 DQ539268 DQ539269 DQ539270 AM765857 AM765868 AM765869 AM765870 AM765871 AM765872 AM765873 AM765874 AM765875 AM765876 ABG02765.1 ABG02766.1 ABG02767.1 ABG02768.1 ABG02769.1 ABG02770.1 ABG02771.1 ABG02772.1 ABG02773.1 ABG02774.1 ABG02775.1 ABG02776.1 ABG02777.1 ABG02779.1 ABG02782.1 ABG02783.1 ABG02784.1 ABG02785.1 ABG02786.1 ABG02787.1 ABG02788.1 ABG02789.1 ABG02790.1 ABG02791.1 CAO78758.1 CAO78769.1 CAO78770.1 CAO78771.1 CAO78772.1 CAO78773.1 CAO78774.1 CAO78775.1 CAO78776.1 CAO78777.1 CM002911 KMY91639.1 AM765859 CAO78760.1 AE013599 AY047548 AAK77280.1 AE014134 AY060796 AAF51277.1 AAL28344.1 KMY91640.1 JX915894 AFZ78860.1 CH480841 EDW49527.1 GEBQ01011698 GEBQ01000257 JAT28279.1 JAT39720.1 AERX01017497 AZIM01002473 ETE63965.1 UFQS01005297 UFQT01005297 SSX16784.1 SSX35971.1 KQ417459 KOF91865.1 REGN01004409 RNA17643.1 CVRI01000038 CRK93881.1 AM765865 CAO78766.1 HADW01014130 SBP15530.1 KQ414637 KOC66990.1
Proteomes
UP000007151
UP000015102
UP000009192
UP000054771
UP000095301
UP000092460
+ More
UP000002640 UP000018468 UP000192221 UP000091820 UP000007798 UP000265100 UP000092443 UP000092444 UP000244855 UP000095300 UP000078200 UP000069272 UP000092445 UP000007801 UP000197619 UP000016666 UP000265160 UP000005207 UP000000539 UP000007267 UP000000803 UP000001292 UP000053454 UP000276133 UP000075884 UP000183832 UP000261520 UP000053825
UP000002640 UP000018468 UP000192221 UP000091820 UP000007798 UP000265100 UP000092443 UP000092444 UP000244855 UP000095300 UP000078200 UP000069272 UP000092445 UP000007801 UP000197619 UP000016666 UP000265160 UP000005207 UP000000539 UP000007267 UP000000803 UP000001292 UP000053454 UP000276133 UP000075884 UP000183832 UP000261520 UP000053825
Pfam
Interpro
IPR009003
Peptidase_S1_PA
+ More
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR001314 Peptidase_S1A
IPR033116 TRYPSIN_SER
IPR040973 CLIP_SPH_Scar
IPR035914 Sperma_CUB_dom_sf
IPR038565 CLIP_sf
IPR022700 CLIP
IPR033040 GZMM
IPR001190 SRCR
IPR000859 CUB_dom
IPR013320 ConA-like_dom_sf
IPR017448 SRCR-like_dom
IPR000998 MAM_dom
IPR023415 LDLR_class-A_CS
IPR036364 SEA_dom_sf
IPR036772 SRCR-like_dom_sf
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR000082 SEA_dom
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR001314 Peptidase_S1A
IPR033116 TRYPSIN_SER
IPR040973 CLIP_SPH_Scar
IPR035914 Sperma_CUB_dom_sf
IPR038565 CLIP_sf
IPR022700 CLIP
IPR033040 GZMM
IPR001190 SRCR
IPR000859 CUB_dom
IPR013320 ConA-like_dom_sf
IPR017448 SRCR-like_dom
IPR000998 MAM_dom
IPR023415 LDLR_class-A_CS
IPR036364 SEA_dom_sf
IPR036772 SRCR-like_dom_sf
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR000082 SEA_dom
SUPFAM
ProteinModelPortal
D9HQ67
D9HQ25
A0A212FKN2
T1GDM2
R4G2C0
R4G7C9
+ More
T1GQB9 B4KXH6 A0A0U5GTC1 A0A1I8MTT1 T1P882 A0A1B0BUI5 G4YJG1 B6Z1X6 A0A1Q1NPF7 W5LWA6 A0A1W4V7B4 A0A1Q1NPF4 A0A1Q1NPI1 A0A1Q1NP77 A0A1W4V5K3 A0A0K8SCH7 A0A146LZD5 A0A034VGE4 A0A0A9W4V0 A0A1A9WYS8 B4N135 A0A034VKN4 A0A3P8NRS8 A0A3P8NRG2 A0A1A9XIV4 A0A1B0FAF0 A0A2V1DTG0 A0A1I8NNQ7 A0A1A9US87 A0A182FUP8 A0A1A9ZAW2 R0JV69 B3MBQ9 A0A218UA67 B4L9R3 T1GQ14 A0A0P9C129 W5LXF7 A7DYW0 A0A310SCG7 Q0GSS5 A7DYV6 Q0GSS6 Q0GSS7 U3IJ08 A0A3P9CA61 A0A0A1XLZ0 A0A2K8JSL6 A0A0K8WBX2 I3K9X1 A0A1D5P7Y3 A7DYV9 A7DYW5 K7FZ69 Q0GST3 Q0GST5 A0A0J9R7H1 A7DYV7 A0A182FUP6 Q7K5M0 Q9VQA0 A0A0J9TTW9 L0ATN6 B4II01 A0A1B6MUX8 I3K8H2 V8NPI1 A0A336LFU1 A0A0L8HT00 A0A3M7R237 A0A182N2U4 A0A1J1I662 A7DYW3 A0A1A7XCF2 A0A3B4AJ83 A0A1A9WAU6 A0A0L7R7Y2
T1GQB9 B4KXH6 A0A0U5GTC1 A0A1I8MTT1 T1P882 A0A1B0BUI5 G4YJG1 B6Z1X6 A0A1Q1NPF7 W5LWA6 A0A1W4V7B4 A0A1Q1NPF4 A0A1Q1NPI1 A0A1Q1NP77 A0A1W4V5K3 A0A0K8SCH7 A0A146LZD5 A0A034VGE4 A0A0A9W4V0 A0A1A9WYS8 B4N135 A0A034VKN4 A0A3P8NRS8 A0A3P8NRG2 A0A1A9XIV4 A0A1B0FAF0 A0A2V1DTG0 A0A1I8NNQ7 A0A1A9US87 A0A182FUP8 A0A1A9ZAW2 R0JV69 B3MBQ9 A0A218UA67 B4L9R3 T1GQ14 A0A0P9C129 W5LXF7 A7DYW0 A0A310SCG7 Q0GSS5 A7DYV6 Q0GSS6 Q0GSS7 U3IJ08 A0A3P9CA61 A0A0A1XLZ0 A0A2K8JSL6 A0A0K8WBX2 I3K9X1 A0A1D5P7Y3 A7DYV9 A7DYW5 K7FZ69 Q0GST3 Q0GST5 A0A0J9R7H1 A7DYV7 A0A182FUP6 Q7K5M0 Q9VQA0 A0A0J9TTW9 L0ATN6 B4II01 A0A1B6MUX8 I3K8H2 V8NPI1 A0A336LFU1 A0A0L8HT00 A0A3M7R237 A0A182N2U4 A0A1J1I662 A7DYW3 A0A1A7XCF2 A0A3B4AJ83 A0A1A9WAU6 A0A0L7R7Y2
PDB
4DGJ
E-value=3.7221e-06,
Score=116
Ontologies
GO
GO:0004252
GO:0046658
GO:0019897
GO:0016021
GO:0008236
GO:0008219
GO:0005044
GO:0016020
GO:0048803
GO:0045197
GO:0055037
GO:0005770
GO:0007390
GO:0090099
GO:0007394
GO:0005615
GO:0016324
GO:0048802
GO:0005769
GO:0006508
GO:0007560
GO:0005737
GO:0050793
GO:0007391
GO:0005576
GO:0004368
GO:0006072
GO:0009331
GO:0005634
GO:0006289
GO:0004518
GO:0006281
GO:0048384
PANTHER
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 20,
Likelihood: 0.979522
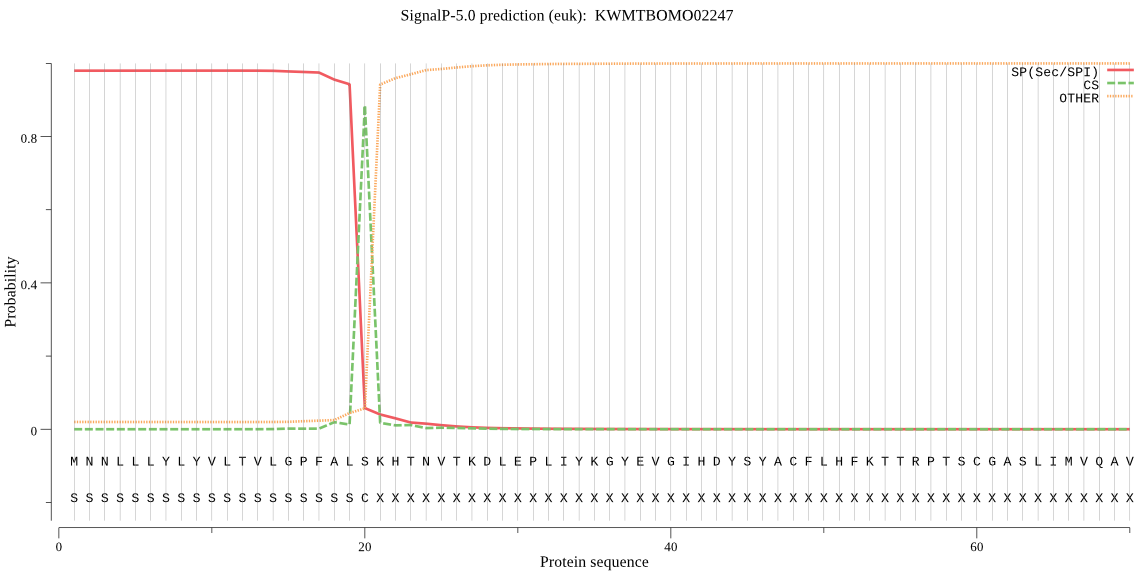
Length:
173
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.21684
Exp number, first 60 AAs:
2.97715
Total prob of N-in:
0.12658
outside
1 - 173
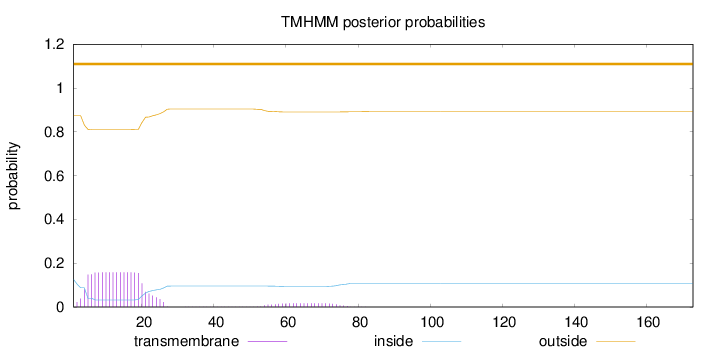
Population Genetic Test Statistics
Pi
134.468775
Theta
151.653333
Tajima's D
-0.35623
CLR
0
CSRT
0.28263586820659
Interpretation
Uncertain
