Gene
KWMTBOMO02242 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003049
Annotation
glycerol-3-phosphate_dehydrogenase_isoform_2_[Bombyx_mori]
Full name
Glycerol-3-phosphate dehydrogenase
Location in the cell
Mitochondrial Reliability : 2.936
Sequence
CDS
ATGTCTAGAAGAAGAATTATTGCAAGTGGAGTAGCTGGCGTTGCGGGTGCAGCCTTAGCCGTGTGGGCCTTCTCCGCCGATGACTCTCCGGTTAACTGGTACTCAGCAGCAACAGTGTCAGCAAAGACCGTACGCAAGAAGCGTCCTCTGCCAACCAGAACAGAACAAGTGGCCAATCTCCAAGCTGGGCACACGTACGATTTGCTCATCATTGGAGGCGGCGCCACGGGAGTTGGGTGTGCACTCGATGCAACAACTAGAGGTCTCCGAACAGCCCTAGTAGAAGCAGACGATTTTGCTAGCGGTACATCCAGTAGAAGCACGAAGCTCATCCATGGCGGAGTCAGGTACCTACAAAAGGCCATCATGCAACTCGACTATGACCAGTACAAGATGGTTAAAGAGGCTCTGCACGAGCGCGCTAATATGCTGGAAGTCGCACCACACTTGACTAGGCCCCTGCCTATTCTTCTACCAGTATACAAATGGTGGCAAGTACCGTATTATTGGTTCGGTATTAAAATGTACGATTTCGTTGCCGGCGACAGAAACGTTAAGAATTCTTATTATTTATCGAAGAAGAATACTCTTGAGTTGTTTCCAATGTTGAAATCGGATAATCTATGCGGAGGAATTGTCTATTATGATGGACAACAAGACGACGCTCGAATGAACTTAGCGATTGCACTGACGGCCGCCCGTCACGGCGCCACCATTGCTAACCACGTCAGCGTCACCAAACTATACAAGGTAGCCGGGAAACTGAGCGGAGCTAGACTGAAGGACGAGTTATCCGGCAAGGAATGGGACGTGCAAGCAAAGTGCATTATCAACGCAACCGGCCCATTCACGGACAGCATCAGGAAAATGGACGATCCCAAAATCAAAGATATCTGCTGTCCATCTTCAGGAGTACATATTGTGTTACCCGGATATTACAGTCCCGAACACATGGGCCTGCTAGACCCGGCAACATCCGACGGCCGCGTCATCTTCTTCTTGCCGTGGCTGAAGGGTACCATCGCCGGAACAACGGACCTGCCCTGCCAAGTCACCCACAACCCTAAACCCACTGAAGACGAAATCATGTTCATCCTTAGCGAAGTCAAGAACTATTTGAATCCGGACGTTGAAGTCCGGCGCGGTGACGTTCTGTCCGCGTGGTCCGGGATCCGGCCGCTAGTGTCGGACCCGAACAAGCCGGACACGCAGTCCCTGGCGCGGAACCACATCGTGCACGTGTCGCCGTCCGGCCTCGTCACCATCGCGGGCGGCAAGTGGACCACGTATCGGTCCATGGCCGCCGAGACCATAGATGCCGCTGTGGAAAGTGCAAATCTCAAACCAATCTTCAGGGACAGTCAGACTGATGGATTCTTAATAGAGGGAGCCCACGGATGGACTCCTACTATGTACATCAGATTGGTGCAAGATTTCGGATTGGAGATGGAAGTCGCCCAGCATTTGTCGAAGTCGTACGGGGACAGAGCGTTTGCTGTTGCCAAGATGGCGACCATGACCGGGAAGAGATGGCCAATCATCGGCAAGAAAATACACCCTGAGTTCCCGTATATAGACGCTGAGATTCGGTATGGAGTCCGTGAATATGCCTGTACAGCTATAGATATGATAGCTAGACGACTTCGTCTCGCCTTCCTCAACGTTCAAGCGGCCGCCGAAGCACTGCCCGTTGTCGTTGACATCATGGCCGAGGAACTCAACTGGAACGAGGCTGAAAGGAAGCGTCAAATTAAAATGGCAAACGAATTCCTTGCAAACGAAATGGGCCAGATGGTGAACCGAGCTAGTCGCGACAAGATCCCCATCAACCTGAGCAAGGAGGAGATACAGACGTACATCAAACGCTTCCAGATCATAGACAAGGATCGGAAGGGATTCGTGTCTATTAACGACATTCGGAGGAGTCTTAAGAACTACGGCGAGGAAGTGACCGGCGAGCAGCTCCACGAGATCCTACGCGAGATCGACACAAACATGAACGGTCAAGTTGAACTCGACGAGTATCTACAGATGATGTCGGCCATTAAATCTGGTCACGTGGCGTACTCGCGTTTCGCTCGAATGGCCGAGATGGAGGAGGAGCATCATGAAAGGGAAACCCTGAAAAAGAAGATCACCGTAGAGAGGAGCGGCGGTGGACTGTAA
Protein
MSRRRIIASGVAGVAGAALAVWAFSADDSPVNWYSAATVSAKTVRKKRPLPTRTEQVANLQAGHTYDLLIIGGGATGVGCALDATTRGLRTALVEADDFASGTSSRSTKLIHGGVRYLQKAIMQLDYDQYKMVKEALHERANMLEVAPHLTRPLPILLPVYKWWQVPYYWFGIKMYDFVAGDRNVKNSYYLSKKNTLELFPMLKSDNLCGGIVYYDGQQDDARMNLAIALTAARHGATIANHVSVTKLYKVAGKLSGARLKDELSGKEWDVQAKCIINATGPFTDSIRKMDDPKIKDICCPSSGVHIVLPGYYSPEHMGLLDPATSDGRVIFFLPWLKGTIAGTTDLPCQVTHNPKPTEDEIMFILSEVKNYLNPDVEVRRGDVLSAWSGIRPLVSDPNKPDTQSLARNHIVHVSPSGLVTIAGGKWTTYRSMAAETIDAAVESANLKPIFRDSQTDGFLIEGAHGWTPTMYIRLVQDFGLEMEVAQHLSKSYGDRAFAVAKMATMTGKRWPIIGKKIHPEFPYIDAEIRYGVREYACTAIDMIARRLRLAFLNVQAAAEALPVVVDIMAEELNWNEAERKRQIKMANEFLANEMGQMVNRASRDKIPINLSKEEIQTYIKRFQIIDKDRKGFVSINDIRRSLKNYGEEVTGEQLHEILREIDTNMNGQVELDEYLQMMSAIKSGHVAYSRFARMAEMEEEHHERETLKKKITVERSGGGL
Summary
Catalytic Activity
a quinone + sn-glycerol 3-phosphate = a quinol + dihydroxyacetone phosphate
Cofactor
FAD
Similarity
Belongs to the FAD-dependent glycerol-3-phosphate dehydrogenase family.
Feature
chain Glycerol-3-phosphate dehydrogenase
Uniprot
C9EJW5
B0EVL7
A0A2A4J8H6
A0A2H1VKH5
A0A2A4J9N7
A0A212F3S6
+ More
A0A194PMD4 A0A2R4SDR8 A0A1B6DUY3 A0A1L8E0W3 A0A310ST21 K7JBG8 A0A0C9RD27 A0A026WSS6 E9ILW1 A0A1L8E0R2 A0A1L8E0R5 B4J7V1 A0A1B6J9Z2 A0A0T6ATN2 A0A1Y1K9X7 A0A1B0CT95 V9IC36 A0A034VQV3 W8BGX8 A0A158NPA2 A0A0M9AA42 A0A034VNY6 A0A1B6E7D0 A0A0L0CBN8 B4H700 Q28WL2 A0A0A1X4X9 A0A067RNM6 B4MJK7 A0A139WHK5 A0A3B0JHL0 A0A195DYA8 A0A0J9RDU7 E0VPS4 B4HSG0 A0A087ZSL0 A0A1W4ULC4 A0A2A3EEM3 T1P9G5 B4P6V2 A0A1I8MW37 B4KRA5 B3NQ13 B3MCF1 A0A0L7QSS0 A0A0M3QVA0 A0A1I8PY97 B4LKQ7 B4QGZ4 Q7K569 D6WMX6 V9IAA7 A0A336MQ85 N6TNJ0 U5EWV1 A0A336MVE8 A0A1J1J686 A0A154PRP5 A0A182JZR5 A0A182MAB2 A0A182Y8Y4 A0A084WP96 A0A182QIQ4 A0A182SXE7 A0A182PLJ4 A0A182VGZ0 A0A182KZR0 F5HLN4 A0A182HYI1 A0A0A9YBB7 F5HLN3 A0A182XF89 A0A182TE52 A0A182RT17 A0A182NL33 B0W992 A0A182W3A7 A0A1Q3FKN3 A0A1Q3FKQ9 A0A182J426 A0A2M4BFD6 A0A182FQ71 A0A182GR19 A0A023EVQ4 A0A2M3ZKN2 A0A2M3YYI4 A0A2M4A6Z9 A0A2M4BF57 E2B8P5 A0A151WXK2 T1DK36 Q17E81 A0A195CIF7 Q17E82
A0A194PMD4 A0A2R4SDR8 A0A1B6DUY3 A0A1L8E0W3 A0A310ST21 K7JBG8 A0A0C9RD27 A0A026WSS6 E9ILW1 A0A1L8E0R2 A0A1L8E0R5 B4J7V1 A0A1B6J9Z2 A0A0T6ATN2 A0A1Y1K9X7 A0A1B0CT95 V9IC36 A0A034VQV3 W8BGX8 A0A158NPA2 A0A0M9AA42 A0A034VNY6 A0A1B6E7D0 A0A0L0CBN8 B4H700 Q28WL2 A0A0A1X4X9 A0A067RNM6 B4MJK7 A0A139WHK5 A0A3B0JHL0 A0A195DYA8 A0A0J9RDU7 E0VPS4 B4HSG0 A0A087ZSL0 A0A1W4ULC4 A0A2A3EEM3 T1P9G5 B4P6V2 A0A1I8MW37 B4KRA5 B3NQ13 B3MCF1 A0A0L7QSS0 A0A0M3QVA0 A0A1I8PY97 B4LKQ7 B4QGZ4 Q7K569 D6WMX6 V9IAA7 A0A336MQ85 N6TNJ0 U5EWV1 A0A336MVE8 A0A1J1J686 A0A154PRP5 A0A182JZR5 A0A182MAB2 A0A182Y8Y4 A0A084WP96 A0A182QIQ4 A0A182SXE7 A0A182PLJ4 A0A182VGZ0 A0A182KZR0 F5HLN4 A0A182HYI1 A0A0A9YBB7 F5HLN3 A0A182XF89 A0A182TE52 A0A182RT17 A0A182NL33 B0W992 A0A182W3A7 A0A1Q3FKN3 A0A1Q3FKQ9 A0A182J426 A0A2M4BFD6 A0A182FQ71 A0A182GR19 A0A023EVQ4 A0A2M3ZKN2 A0A2M3YYI4 A0A2M4A6Z9 A0A2M4BF57 E2B8P5 A0A151WXK2 T1DK36 Q17E81 A0A195CIF7 Q17E82
EC Number
1.1.5.3
Pubmed
19121390
22118469
26354079
20075255
24508170
30249741
+ More
21282665 17994087 28004739 25348373 24495485 21347285 26108605 15632085 23185243 25830018 24845553 18362917 19820115 22936249 20566863 17550304 25315136 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 25244985 24438588 20966253 12364791 14747013 17210077 25401762 26823975 26483478 24945155 20798317 17510324
21282665 17994087 28004739 25348373 24495485 21347285 26108605 15632085 23185243 25830018 24845553 18362917 19820115 22936249 20566863 17550304 25315136 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 25244985 24438588 20966253 12364791 14747013 17210077 25401762 26823975 26483478 24945155 20798317 17510324
EMBL
GQ865685
GQ865686
ACV95334.1
BABH01010255
BABH01010256
BABH01010257
+ More
EF154335 GQ865687 GQ865688 ABM46603.2 ACV95336.1 NWSH01002494 PCG68169.1 ODYU01003054 SOQ41349.1 PCG68170.1 AGBW02010507 OWR48372.1 KQ459598 KPI94596.1 MF101766 AVZ46903.1 GEDC01027407 GEDC01007815 JAS09891.1 JAS29483.1 GFDF01001765 JAV12319.1 KQ760505 OAD60070.1 AAZX01012716 GBYB01011047 JAG80814.1 KK107119 QOIP01000009 EZA58721.1 RLU18934.1 GL764129 EFZ18296.1 GFDF01001766 JAV12318.1 GFDF01001764 JAV12320.1 CH916367 EDW01157.1 GECU01018991 GECU01011679 GECU01006943 GECU01005602 JAS88715.1 JAS96027.1 JAT00764.1 JAT02105.1 LJIG01022836 KRT78466.1 GEZM01088494 GEZM01088493 JAV58164.1 AJWK01027296 AJWK01027297 AJWK01027298 AJWK01027299 JR037925 AEY58036.1 GAKP01014782 GAKP01014778 JAC44170.1 GAMC01006035 JAC00521.1 ADTU01022311 KQ435718 KOX78959.1 GAKP01014783 GAKP01014779 GAKP01014775 GAKP01014774 GAKP01014772 JAC44169.1 GEDC01011959 GEDC01003457 JAS25339.1 JAS33841.1 JRES01000644 KNC29671.1 CH479216 EDW33636.1 CM000071 EAL26655.1 KRT03276.1 GBXI01008110 JAD06182.1 KK852561 KDR21344.1 CH963846 EDW72296.1 KQ971343 KYB27335.1 OUUW01000001 SPP72726.1 KQ980066 KYN17895.1 CM002911 KMY94096.1 DS235371 EEB15380.1 CH480816 EDW48038.1 KZ288266 PBC30205.1 KA644553 AFP59182.1 CM000158 EDW92029.1 CH933808 EDW09321.1 CH954179 EDV55860.1 CH902619 EDV37275.1 KPU76675.1 KQ414756 KOC61599.1 CP012524 ALC42013.1 CH940648 EDW61780.1 CM000362 EDX07257.1 AE013599 AY051430 AAF58099.2 AAK92854.1 EFA04316.2 JR037924 AEY58035.1 UFQS01002125 UFQT01002125 SSX13277.1 SSX32714.1 APGK01028342 APGK01028343 APGK01028344 APGK01028345 KB740686 KB631930 ENN79553.1 ERL87310.1 GANO01001254 JAB58617.1 SSX13276.1 SSX32713.1 CVRI01000074 CRL07917.1 KQ435090 KZC14579.1 AXCM01001209 ATLV01024994 KE525367 KFB52040.1 AXCN02000087 AAAB01008898 EGK97166.1 APCN01005372 GBHO01013227 GBRD01010063 GDHC01000856 JAG30377.1 JAG55761.1 JAQ17773.1 EGK97167.1 DS231862 EDS39789.1 GFDL01006927 JAV28118.1 GFDL01006930 JAV28115.1 GGFJ01002562 MBW51703.1 JXUM01082801 KQ563337 KXJ74027.1 GAPW01000330 JAC13268.1 GGFM01008343 MBW29094.1 GGFM01000565 MBW21316.1 GGFK01003243 MBW36564.1 GGFJ01002544 MBW51685.1 GL446361 EFN87951.1 KQ982668 KYQ52497.1 GAMD01001146 JAB00445.1 CH477284 EAT44796.1 KQ977791 KYM99848.1 EAT44797.1
EF154335 GQ865687 GQ865688 ABM46603.2 ACV95336.1 NWSH01002494 PCG68169.1 ODYU01003054 SOQ41349.1 PCG68170.1 AGBW02010507 OWR48372.1 KQ459598 KPI94596.1 MF101766 AVZ46903.1 GEDC01027407 GEDC01007815 JAS09891.1 JAS29483.1 GFDF01001765 JAV12319.1 KQ760505 OAD60070.1 AAZX01012716 GBYB01011047 JAG80814.1 KK107119 QOIP01000009 EZA58721.1 RLU18934.1 GL764129 EFZ18296.1 GFDF01001766 JAV12318.1 GFDF01001764 JAV12320.1 CH916367 EDW01157.1 GECU01018991 GECU01011679 GECU01006943 GECU01005602 JAS88715.1 JAS96027.1 JAT00764.1 JAT02105.1 LJIG01022836 KRT78466.1 GEZM01088494 GEZM01088493 JAV58164.1 AJWK01027296 AJWK01027297 AJWK01027298 AJWK01027299 JR037925 AEY58036.1 GAKP01014782 GAKP01014778 JAC44170.1 GAMC01006035 JAC00521.1 ADTU01022311 KQ435718 KOX78959.1 GAKP01014783 GAKP01014779 GAKP01014775 GAKP01014774 GAKP01014772 JAC44169.1 GEDC01011959 GEDC01003457 JAS25339.1 JAS33841.1 JRES01000644 KNC29671.1 CH479216 EDW33636.1 CM000071 EAL26655.1 KRT03276.1 GBXI01008110 JAD06182.1 KK852561 KDR21344.1 CH963846 EDW72296.1 KQ971343 KYB27335.1 OUUW01000001 SPP72726.1 KQ980066 KYN17895.1 CM002911 KMY94096.1 DS235371 EEB15380.1 CH480816 EDW48038.1 KZ288266 PBC30205.1 KA644553 AFP59182.1 CM000158 EDW92029.1 CH933808 EDW09321.1 CH954179 EDV55860.1 CH902619 EDV37275.1 KPU76675.1 KQ414756 KOC61599.1 CP012524 ALC42013.1 CH940648 EDW61780.1 CM000362 EDX07257.1 AE013599 AY051430 AAF58099.2 AAK92854.1 EFA04316.2 JR037924 AEY58035.1 UFQS01002125 UFQT01002125 SSX13277.1 SSX32714.1 APGK01028342 APGK01028343 APGK01028344 APGK01028345 KB740686 KB631930 ENN79553.1 ERL87310.1 GANO01001254 JAB58617.1 SSX13276.1 SSX32713.1 CVRI01000074 CRL07917.1 KQ435090 KZC14579.1 AXCM01001209 ATLV01024994 KE525367 KFB52040.1 AXCN02000087 AAAB01008898 EGK97166.1 APCN01005372 GBHO01013227 GBRD01010063 GDHC01000856 JAG30377.1 JAG55761.1 JAQ17773.1 EGK97167.1 DS231862 EDS39789.1 GFDL01006927 JAV28118.1 GFDL01006930 JAV28115.1 GGFJ01002562 MBW51703.1 JXUM01082801 KQ563337 KXJ74027.1 GAPW01000330 JAC13268.1 GGFM01008343 MBW29094.1 GGFM01000565 MBW21316.1 GGFK01003243 MBW36564.1 GGFJ01002544 MBW51685.1 GL446361 EFN87951.1 KQ982668 KYQ52497.1 GAMD01001146 JAB00445.1 CH477284 EAT44796.1 KQ977791 KYM99848.1 EAT44797.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000002358
UP000053097
+ More
UP000279307 UP000001070 UP000092461 UP000005205 UP000053105 UP000037069 UP000008744 UP000001819 UP000027135 UP000007798 UP000007266 UP000268350 UP000078492 UP000009046 UP000001292 UP000005203 UP000192221 UP000242457 UP000002282 UP000095301 UP000009192 UP000008711 UP000007801 UP000053825 UP000092553 UP000095300 UP000008792 UP000000304 UP000000803 UP000019118 UP000030742 UP000183832 UP000076502 UP000075881 UP000075883 UP000076408 UP000030765 UP000075886 UP000075901 UP000075885 UP000075903 UP000075882 UP000007062 UP000075840 UP000076407 UP000075902 UP000075900 UP000075884 UP000002320 UP000075920 UP000075880 UP000069272 UP000069940 UP000249989 UP000008237 UP000075809 UP000008820 UP000078542
UP000279307 UP000001070 UP000092461 UP000005205 UP000053105 UP000037069 UP000008744 UP000001819 UP000027135 UP000007798 UP000007266 UP000268350 UP000078492 UP000009046 UP000001292 UP000005203 UP000192221 UP000242457 UP000002282 UP000095301 UP000009192 UP000008711 UP000007801 UP000053825 UP000092553 UP000095300 UP000008792 UP000000304 UP000000803 UP000019118 UP000030742 UP000183832 UP000076502 UP000075881 UP000075883 UP000076408 UP000030765 UP000075886 UP000075901 UP000075885 UP000075903 UP000075882 UP000007062 UP000075840 UP000076407 UP000075902 UP000075900 UP000075884 UP000002320 UP000075920 UP000075880 UP000069272 UP000069940 UP000249989 UP000008237 UP000075809 UP000008820 UP000078542
Interpro
Gene 3D
CDD
ProteinModelPortal
C9EJW5
B0EVL7
A0A2A4J8H6
A0A2H1VKH5
A0A2A4J9N7
A0A212F3S6
+ More
A0A194PMD4 A0A2R4SDR8 A0A1B6DUY3 A0A1L8E0W3 A0A310ST21 K7JBG8 A0A0C9RD27 A0A026WSS6 E9ILW1 A0A1L8E0R2 A0A1L8E0R5 B4J7V1 A0A1B6J9Z2 A0A0T6ATN2 A0A1Y1K9X7 A0A1B0CT95 V9IC36 A0A034VQV3 W8BGX8 A0A158NPA2 A0A0M9AA42 A0A034VNY6 A0A1B6E7D0 A0A0L0CBN8 B4H700 Q28WL2 A0A0A1X4X9 A0A067RNM6 B4MJK7 A0A139WHK5 A0A3B0JHL0 A0A195DYA8 A0A0J9RDU7 E0VPS4 B4HSG0 A0A087ZSL0 A0A1W4ULC4 A0A2A3EEM3 T1P9G5 B4P6V2 A0A1I8MW37 B4KRA5 B3NQ13 B3MCF1 A0A0L7QSS0 A0A0M3QVA0 A0A1I8PY97 B4LKQ7 B4QGZ4 Q7K569 D6WMX6 V9IAA7 A0A336MQ85 N6TNJ0 U5EWV1 A0A336MVE8 A0A1J1J686 A0A154PRP5 A0A182JZR5 A0A182MAB2 A0A182Y8Y4 A0A084WP96 A0A182QIQ4 A0A182SXE7 A0A182PLJ4 A0A182VGZ0 A0A182KZR0 F5HLN4 A0A182HYI1 A0A0A9YBB7 F5HLN3 A0A182XF89 A0A182TE52 A0A182RT17 A0A182NL33 B0W992 A0A182W3A7 A0A1Q3FKN3 A0A1Q3FKQ9 A0A182J426 A0A2M4BFD6 A0A182FQ71 A0A182GR19 A0A023EVQ4 A0A2M3ZKN2 A0A2M3YYI4 A0A2M4A6Z9 A0A2M4BF57 E2B8P5 A0A151WXK2 T1DK36 Q17E81 A0A195CIF7 Q17E82
A0A194PMD4 A0A2R4SDR8 A0A1B6DUY3 A0A1L8E0W3 A0A310ST21 K7JBG8 A0A0C9RD27 A0A026WSS6 E9ILW1 A0A1L8E0R2 A0A1L8E0R5 B4J7V1 A0A1B6J9Z2 A0A0T6ATN2 A0A1Y1K9X7 A0A1B0CT95 V9IC36 A0A034VQV3 W8BGX8 A0A158NPA2 A0A0M9AA42 A0A034VNY6 A0A1B6E7D0 A0A0L0CBN8 B4H700 Q28WL2 A0A0A1X4X9 A0A067RNM6 B4MJK7 A0A139WHK5 A0A3B0JHL0 A0A195DYA8 A0A0J9RDU7 E0VPS4 B4HSG0 A0A087ZSL0 A0A1W4ULC4 A0A2A3EEM3 T1P9G5 B4P6V2 A0A1I8MW37 B4KRA5 B3NQ13 B3MCF1 A0A0L7QSS0 A0A0M3QVA0 A0A1I8PY97 B4LKQ7 B4QGZ4 Q7K569 D6WMX6 V9IAA7 A0A336MQ85 N6TNJ0 U5EWV1 A0A336MVE8 A0A1J1J686 A0A154PRP5 A0A182JZR5 A0A182MAB2 A0A182Y8Y4 A0A084WP96 A0A182QIQ4 A0A182SXE7 A0A182PLJ4 A0A182VGZ0 A0A182KZR0 F5HLN4 A0A182HYI1 A0A0A9YBB7 F5HLN3 A0A182XF89 A0A182TE52 A0A182RT17 A0A182NL33 B0W992 A0A182W3A7 A0A1Q3FKN3 A0A1Q3FKQ9 A0A182J426 A0A2M4BFD6 A0A182FQ71 A0A182GR19 A0A023EVQ4 A0A2M3ZKN2 A0A2M3YYI4 A0A2M4A6Z9 A0A2M4BF57 E2B8P5 A0A151WXK2 T1DK36 Q17E81 A0A195CIF7 Q17E82
PDB
3DA1
E-value=1.34895e-70,
Score=679
Ontologies
GO
PANTHER
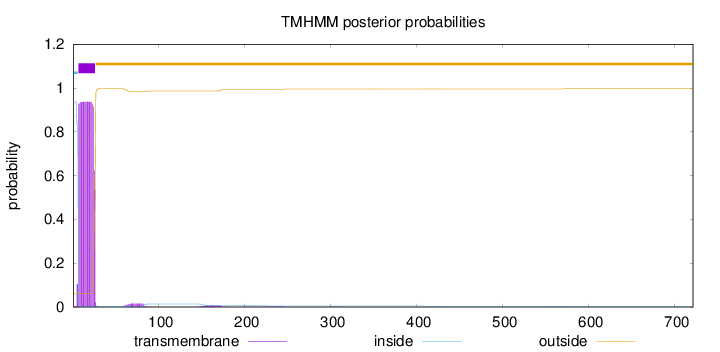
Topology
Length:
721
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.7717199999999
Exp number, first 60 AAs:
18.2224
Total prob of N-in:
0.93691
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 26
outside
27 - 721
Population Genetic Test Statistics
Pi
217.159985
Theta
155.465668
Tajima's D
1.271299
CLR
0.826267
CSRT
0.728163591820409
Interpretation
Uncertain
