Pre Gene Modal
BGIBMGA003048
Annotation
signal_peptidase_complex_subunit_2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.231
Sequence
CDS
ATGTCTGAAACGGCGGAGGCTGCAAAGATCAACAAATGGGATGGTGCTGCAGCAAAAAATGCTGTTGATGATGCTATTAGAGAGGTAATGACTGGAGACTTAAAATGCAAGGAAAGCTTTGCTCTAATTGACGGTAGACTGTTCCTCTGTGCTCTGGCTGTTGGTGTGGCCCTTTACGCACTGCTATGGGATTATCTTTATCCATTCCCTCAATCAAGACTGGTTCTAATCATCTGTGTGTCATCATATTTCATACTGATGGGTATTTTGACCCTCTACACTACATTCAAAGAGAAGGGCATCTTTGTGGTAGCCAAGGAGAAGGTTGGAAATAACACCAGGGTCTGGGAAGCCAGTTCTTATGTAAAGAAACACGACGATAAATACAATCTCGTAATTGTTATGCGAGACACGAATGGCAACACTCGTGAGGCATCCGTGACCAAGTCGTTTGCCAACTTCATCGACGTGAACGGCACTGTAGTTCAAAACATTGTATCTAATGAGATCACCAAGCTGTACCACAGCCTCTCATCTGAGAAGAAGGAAAAGTAA
Protein
MSETAEAAKINKWDGAAAKNAVDDAIREVMTGDLKCKESFALIDGRLFLCALAVGVALYALLWDYLYPFPQSRLVLIICVSSYFILMGILTLYTTFKEKGIFVVAKEKVGNNTRVWEASSYVKKHDDKYNLVIVMRDTNGNTREASVTKSFANFIDVNGTVVQNIVSNEITKLYHSLSSEKKEK
Summary
Uniprot
Q2F5W0
H9J0L1
A0A0N1I6L5
A0A2H1V606
A0A2A4J912
A0A1E1WQ68
+ More
A0A212F3T8 I4DJH2 S4PBB5 A0A1B0D490 A0A1L8DUH1 A0A1Y1K8E3 A0A1B0GI44 A0A1W4XDY3 A0A2P8XLC6 A0A0K8TSU6 T1HL74 R4G567 A0A224XL66 V5FZ74 G3CJT3 A0A067QQ52 A0A023F602 R4WSF8 A0A336MPF4 A0A1B6HWM9 A0A1B6FFI2 E2BWB2 A0A0N0BIM0 A0A1B6L2K1 A0A088AHR3 E2AIH6 A0A158NGX9 A0A195B6L5 A0A195FN76 A0A2A3EET1 V9IGX5 A0A3L8DQ93 D6WNW8 A0A0A9WHH1 N6U0A5 D1FPM8 A0A1B6BZ02 A0A182T6K8 A0A182W1Z0 A0A182Y9C2 A0A0L7RHU1 A0A182R8T8 A0A2M4C088 A0A182MBK1 A0A1S3D4N2 A0A2M4A7T8 A0A182N3X7 A0A182FUY8 B4L1K1 A0A154P7V4 A0A182Q0S9 W5JVD1 E0VF85 A0A2M3ZG98 T1EAS1 A0A0J7KDR0 U5ESF7 A0A182K6W9 A0A182P4Z8 A0A1J1IP71 B4M358 B3MAM6 A0A232EQP6 A0A0C9S1W8 A0A034WEN3 A0A0L0BTN7 B0WS21 A0A182VJV3 A0A1I8PZ10 A0A3B0JXT3 A0A182XGT7 Q7PZC7 A0A182HHF4 W8AAK6 A0A023EIM2 C6ZQT3 A0A1Q3FHF6 Q29HS9 A0A0P6GY50 A0A182UGQ4 A0A3B0KM13 A0A1W4UNV1 Q1HRP4 A0A226DNK3 A0A0P6G2H9 A0A1L8EFM3 A0A0P6H7J2 A0A1A9XXQ6 A0A1A9WZC0 A0A1L8EFR9 A0A1B0APG9 A0A162BP07 A0A0P5BJZ5 A0A0P5UDU0
A0A212F3T8 I4DJH2 S4PBB5 A0A1B0D490 A0A1L8DUH1 A0A1Y1K8E3 A0A1B0GI44 A0A1W4XDY3 A0A2P8XLC6 A0A0K8TSU6 T1HL74 R4G567 A0A224XL66 V5FZ74 G3CJT3 A0A067QQ52 A0A023F602 R4WSF8 A0A336MPF4 A0A1B6HWM9 A0A1B6FFI2 E2BWB2 A0A0N0BIM0 A0A1B6L2K1 A0A088AHR3 E2AIH6 A0A158NGX9 A0A195B6L5 A0A195FN76 A0A2A3EET1 V9IGX5 A0A3L8DQ93 D6WNW8 A0A0A9WHH1 N6U0A5 D1FPM8 A0A1B6BZ02 A0A182T6K8 A0A182W1Z0 A0A182Y9C2 A0A0L7RHU1 A0A182R8T8 A0A2M4C088 A0A182MBK1 A0A1S3D4N2 A0A2M4A7T8 A0A182N3X7 A0A182FUY8 B4L1K1 A0A154P7V4 A0A182Q0S9 W5JVD1 E0VF85 A0A2M3ZG98 T1EAS1 A0A0J7KDR0 U5ESF7 A0A182K6W9 A0A182P4Z8 A0A1J1IP71 B4M358 B3MAM6 A0A232EQP6 A0A0C9S1W8 A0A034WEN3 A0A0L0BTN7 B0WS21 A0A182VJV3 A0A1I8PZ10 A0A3B0JXT3 A0A182XGT7 Q7PZC7 A0A182HHF4 W8AAK6 A0A023EIM2 C6ZQT3 A0A1Q3FHF6 Q29HS9 A0A0P6GY50 A0A182UGQ4 A0A3B0KM13 A0A1W4UNV1 Q1HRP4 A0A226DNK3 A0A0P6G2H9 A0A1L8EFM3 A0A0P6H7J2 A0A1A9XXQ6 A0A1A9WZC0 A0A1L8EFR9 A0A1B0APG9 A0A162BP07 A0A0P5BJZ5 A0A0P5UDU0
Pubmed
19121390
26354079
22118469
22651552
23622113
28004739
+ More
29403074 26369729 21058630 24845553 25474469 23691247 20798317 21347285 30249741 18362917 19820115 25401762 26823975 23537049 20441151 25244985 17994087 20920257 23761445 20566863 28648823 25348373 26108605 12364791 14747013 17210077 24495485 24945155 26483478 20496585 15632085 17204158 17510324
29403074 26369729 21058630 24845553 25474469 23691247 20798317 21347285 30249741 18362917 19820115 25401762 26823975 23537049 20441151 25244985 17994087 20920257 23761445 20566863 28648823 25348373 26108605 12364791 14747013 17210077 24495485 24945155 26483478 20496585 15632085 17204158 17510324
EMBL
DQ311313
ABD36257.1
BABH01010257
KQ461183
KPJ07565.1
ODYU01000860
+ More
SOQ36261.1 NWSH01002494 PCG68168.1 GDQN01001895 JAT89159.1 AGBW02010507 OWR48373.1 AK401440 KQ459598 BAM18062.1 KPI94597.1 GAIX01005992 JAA86568.1 AJVK01011289 GFDF01004001 JAV10083.1 GEZM01092764 JAV56470.1 AJWK01011279 PYGN01001791 PSN32797.1 GDAI01000402 JAI17201.1 ACPB03006655 GAHY01000176 JAA77334.1 GFTR01003211 JAW13215.1 GALX01005476 JAB62990.1 HP639850 AEM97987.1 KK853149 KDR10695.1 GBBI01002085 JAC16627.1 AK417637 BAN20852.1 UFQS01001975 UFQT01001975 SSX12830.1 SSX32272.1 GECU01028671 JAS79035.1 GECZ01025773 GECZ01020812 JAS43996.1 JAS48957.1 GL451103 EFN80016.1 KQ435727 KOX77868.1 GEBQ01022024 JAT17953.1 GL439791 EFN66766.1 ADTU01015319 KQ976574 KYM80158.1 KQ981387 KYN42030.1 KZ288282 PBC29521.1 JR047077 AEY60320.1 QOIP01000005 RLU22541.1 KQ971343 EFA03199.1 GBHO01037646 GBHO01037644 GBHO01037643 GBHO01037640 GBRD01015528 GDHC01010946 GDHC01005394 JAG05958.1 JAG05960.1 JAG05961.1 JAG05964.1 JAG50298.1 JAQ07683.1 JAQ13235.1 APGK01054307 KB741248 KB631667 ENN71972.1 ERL85151.1 EZ419778 ACY69951.1 GEDC01031014 JAS06284.1 KQ414584 KOC70557.1 GGFJ01009598 MBW58739.1 AXCM01007101 GGFK01003535 MBW36856.1 CH933810 EDW07637.1 KQ434826 KZC07414.1 AXCN02001095 ADMH02000001 ETN68146.1 DS235107 EEB12041.1 GGFM01006788 MBW27539.1 GAMD01000793 JAB00798.1 LBMM01009164 KMQ88336.1 GANO01002377 JAB57494.1 CVRI01000054 CRL00289.1 CH940651 EDW65233.1 CH902618 EDV41313.1 NNAY01002733 OXU20668.1 GBYB01014811 JAG84578.1 GAKP01006196 GAKP01006195 JAC52757.1 JRES01001480 KNC22594.1 DS232063 EDS33639.1 OUUW01000011 SPP86875.1 AAAB01008986 EAA00503.4 APCN01002408 GAMC01021665 JAB84890.1 JXUM01051027 GAPW01004662 GAPW01004661 GAPW01004660 GAPW01004659 KQ561672 JAC08937.1 KXJ77845.1 EZ114884 ACU30937.1 GFDL01008071 JAV26974.1 CH379064 EAL31678.2 GDIQ01039441 JAN55296.1 SPP86876.1 DQ440050 CH477271 ABF18083.1 EAT45290.1 LNIX01000015 OXA46424.1 GDIQ01039440 JAN55297.1 GFDG01001384 JAV17415.1 GDIQ01023138 JAN71599.1 GFDG01001386 JAV17413.1 JXJN01001399 LRGB01027721 KZR95670.1 GDIP01183503 JAJ39899.1 GDIP01237002 GDIP01118209 JAL85505.1
SOQ36261.1 NWSH01002494 PCG68168.1 GDQN01001895 JAT89159.1 AGBW02010507 OWR48373.1 AK401440 KQ459598 BAM18062.1 KPI94597.1 GAIX01005992 JAA86568.1 AJVK01011289 GFDF01004001 JAV10083.1 GEZM01092764 JAV56470.1 AJWK01011279 PYGN01001791 PSN32797.1 GDAI01000402 JAI17201.1 ACPB03006655 GAHY01000176 JAA77334.1 GFTR01003211 JAW13215.1 GALX01005476 JAB62990.1 HP639850 AEM97987.1 KK853149 KDR10695.1 GBBI01002085 JAC16627.1 AK417637 BAN20852.1 UFQS01001975 UFQT01001975 SSX12830.1 SSX32272.1 GECU01028671 JAS79035.1 GECZ01025773 GECZ01020812 JAS43996.1 JAS48957.1 GL451103 EFN80016.1 KQ435727 KOX77868.1 GEBQ01022024 JAT17953.1 GL439791 EFN66766.1 ADTU01015319 KQ976574 KYM80158.1 KQ981387 KYN42030.1 KZ288282 PBC29521.1 JR047077 AEY60320.1 QOIP01000005 RLU22541.1 KQ971343 EFA03199.1 GBHO01037646 GBHO01037644 GBHO01037643 GBHO01037640 GBRD01015528 GDHC01010946 GDHC01005394 JAG05958.1 JAG05960.1 JAG05961.1 JAG05964.1 JAG50298.1 JAQ07683.1 JAQ13235.1 APGK01054307 KB741248 KB631667 ENN71972.1 ERL85151.1 EZ419778 ACY69951.1 GEDC01031014 JAS06284.1 KQ414584 KOC70557.1 GGFJ01009598 MBW58739.1 AXCM01007101 GGFK01003535 MBW36856.1 CH933810 EDW07637.1 KQ434826 KZC07414.1 AXCN02001095 ADMH02000001 ETN68146.1 DS235107 EEB12041.1 GGFM01006788 MBW27539.1 GAMD01000793 JAB00798.1 LBMM01009164 KMQ88336.1 GANO01002377 JAB57494.1 CVRI01000054 CRL00289.1 CH940651 EDW65233.1 CH902618 EDV41313.1 NNAY01002733 OXU20668.1 GBYB01014811 JAG84578.1 GAKP01006196 GAKP01006195 JAC52757.1 JRES01001480 KNC22594.1 DS232063 EDS33639.1 OUUW01000011 SPP86875.1 AAAB01008986 EAA00503.4 APCN01002408 GAMC01021665 JAB84890.1 JXUM01051027 GAPW01004662 GAPW01004661 GAPW01004660 GAPW01004659 KQ561672 JAC08937.1 KXJ77845.1 EZ114884 ACU30937.1 GFDL01008071 JAV26974.1 CH379064 EAL31678.2 GDIQ01039441 JAN55296.1 SPP86876.1 DQ440050 CH477271 ABF18083.1 EAT45290.1 LNIX01000015 OXA46424.1 GDIQ01039440 JAN55297.1 GFDG01001384 JAV17415.1 GDIQ01023138 JAN71599.1 GFDG01001386 JAV17413.1 JXJN01001399 LRGB01027721 KZR95670.1 GDIP01183503 JAJ39899.1 GDIP01237002 GDIP01118209 JAL85505.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000053268
UP000092462
+ More
UP000092461 UP000192223 UP000245037 UP000015103 UP000027135 UP000008237 UP000053105 UP000005203 UP000000311 UP000005205 UP000078540 UP000078541 UP000242457 UP000279307 UP000007266 UP000019118 UP000030742 UP000075901 UP000075920 UP000076408 UP000053825 UP000075900 UP000075883 UP000079169 UP000075884 UP000069272 UP000009192 UP000076502 UP000075886 UP000000673 UP000009046 UP000036403 UP000075881 UP000075885 UP000183832 UP000008792 UP000007801 UP000215335 UP000037069 UP000002320 UP000075903 UP000095300 UP000268350 UP000076407 UP000007062 UP000075840 UP000069940 UP000249989 UP000001819 UP000075902 UP000192221 UP000008820 UP000198287 UP000092443 UP000091820 UP000092460 UP000076858
UP000092461 UP000192223 UP000245037 UP000015103 UP000027135 UP000008237 UP000053105 UP000005203 UP000000311 UP000005205 UP000078540 UP000078541 UP000242457 UP000279307 UP000007266 UP000019118 UP000030742 UP000075901 UP000075920 UP000076408 UP000053825 UP000075900 UP000075883 UP000079169 UP000075884 UP000069272 UP000009192 UP000076502 UP000075886 UP000000673 UP000009046 UP000036403 UP000075881 UP000075885 UP000183832 UP000008792 UP000007801 UP000215335 UP000037069 UP000002320 UP000075903 UP000095300 UP000268350 UP000076407 UP000007062 UP000075840 UP000069940 UP000249989 UP000001819 UP000075902 UP000192221 UP000008820 UP000198287 UP000092443 UP000091820 UP000092460 UP000076858
Pfam
PF06703 SPC25
ProteinModelPortal
Q2F5W0
H9J0L1
A0A0N1I6L5
A0A2H1V606
A0A2A4J912
A0A1E1WQ68
+ More
A0A212F3T8 I4DJH2 S4PBB5 A0A1B0D490 A0A1L8DUH1 A0A1Y1K8E3 A0A1B0GI44 A0A1W4XDY3 A0A2P8XLC6 A0A0K8TSU6 T1HL74 R4G567 A0A224XL66 V5FZ74 G3CJT3 A0A067QQ52 A0A023F602 R4WSF8 A0A336MPF4 A0A1B6HWM9 A0A1B6FFI2 E2BWB2 A0A0N0BIM0 A0A1B6L2K1 A0A088AHR3 E2AIH6 A0A158NGX9 A0A195B6L5 A0A195FN76 A0A2A3EET1 V9IGX5 A0A3L8DQ93 D6WNW8 A0A0A9WHH1 N6U0A5 D1FPM8 A0A1B6BZ02 A0A182T6K8 A0A182W1Z0 A0A182Y9C2 A0A0L7RHU1 A0A182R8T8 A0A2M4C088 A0A182MBK1 A0A1S3D4N2 A0A2M4A7T8 A0A182N3X7 A0A182FUY8 B4L1K1 A0A154P7V4 A0A182Q0S9 W5JVD1 E0VF85 A0A2M3ZG98 T1EAS1 A0A0J7KDR0 U5ESF7 A0A182K6W9 A0A182P4Z8 A0A1J1IP71 B4M358 B3MAM6 A0A232EQP6 A0A0C9S1W8 A0A034WEN3 A0A0L0BTN7 B0WS21 A0A182VJV3 A0A1I8PZ10 A0A3B0JXT3 A0A182XGT7 Q7PZC7 A0A182HHF4 W8AAK6 A0A023EIM2 C6ZQT3 A0A1Q3FHF6 Q29HS9 A0A0P6GY50 A0A182UGQ4 A0A3B0KM13 A0A1W4UNV1 Q1HRP4 A0A226DNK3 A0A0P6G2H9 A0A1L8EFM3 A0A0P6H7J2 A0A1A9XXQ6 A0A1A9WZC0 A0A1L8EFR9 A0A1B0APG9 A0A162BP07 A0A0P5BJZ5 A0A0P5UDU0
A0A212F3T8 I4DJH2 S4PBB5 A0A1B0D490 A0A1L8DUH1 A0A1Y1K8E3 A0A1B0GI44 A0A1W4XDY3 A0A2P8XLC6 A0A0K8TSU6 T1HL74 R4G567 A0A224XL66 V5FZ74 G3CJT3 A0A067QQ52 A0A023F602 R4WSF8 A0A336MPF4 A0A1B6HWM9 A0A1B6FFI2 E2BWB2 A0A0N0BIM0 A0A1B6L2K1 A0A088AHR3 E2AIH6 A0A158NGX9 A0A195B6L5 A0A195FN76 A0A2A3EET1 V9IGX5 A0A3L8DQ93 D6WNW8 A0A0A9WHH1 N6U0A5 D1FPM8 A0A1B6BZ02 A0A182T6K8 A0A182W1Z0 A0A182Y9C2 A0A0L7RHU1 A0A182R8T8 A0A2M4C088 A0A182MBK1 A0A1S3D4N2 A0A2M4A7T8 A0A182N3X7 A0A182FUY8 B4L1K1 A0A154P7V4 A0A182Q0S9 W5JVD1 E0VF85 A0A2M3ZG98 T1EAS1 A0A0J7KDR0 U5ESF7 A0A182K6W9 A0A182P4Z8 A0A1J1IP71 B4M358 B3MAM6 A0A232EQP6 A0A0C9S1W8 A0A034WEN3 A0A0L0BTN7 B0WS21 A0A182VJV3 A0A1I8PZ10 A0A3B0JXT3 A0A182XGT7 Q7PZC7 A0A182HHF4 W8AAK6 A0A023EIM2 C6ZQT3 A0A1Q3FHF6 Q29HS9 A0A0P6GY50 A0A182UGQ4 A0A3B0KM13 A0A1W4UNV1 Q1HRP4 A0A226DNK3 A0A0P6G2H9 A0A1L8EFM3 A0A0P6H7J2 A0A1A9XXQ6 A0A1A9WZC0 A0A1L8EFR9 A0A1B0APG9 A0A162BP07 A0A0P5BJZ5 A0A0P5UDU0
Ontologies
GO
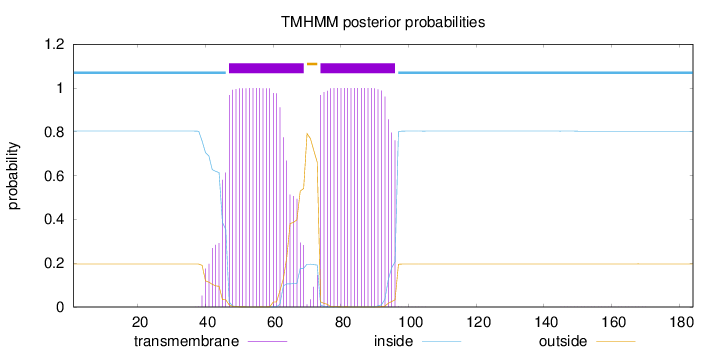
Topology
Length:
184
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.40982
Exp number, first 60 AAs:
16.387
Total prob of N-in:
0.80408
POSSIBLE N-term signal
sequence
inside
1 - 46
TMhelix
47 - 69
outside
70 - 73
TMhelix
74 - 96
inside
97 - 184
Population Genetic Test Statistics
Pi
202.717474
Theta
173.026833
Tajima's D
0.702652
CLR
0
CSRT
0.574571271436428
Interpretation
Uncertain
