Gene
KWMTBOMO02239
Pre Gene Modal
BGIBMGA003107
Annotation
PREDICTED:_histone-lysine_N-methyltransferase_SETD2_isoform_X2_[Bombyx_mori]
Full name
Histone-lysine N-methyltransferase
Location in the cell
Nuclear Reliability : 4.193
Sequence
CDS
ATGTCAGTCGAGGACAGTATAAATGATAGAAACTTTTTATTCCAAGCAGAGGACAGTTCATCGGACGAAGAGGAGCAATTGAACAGCTCTTTGGACAGTCCGCTGGTGATGCGACATGCTAAAGGGAAACTTGTTGAAGGGGTCAATGGTATATATGAAGTAATCAAAGAAGATGCTCAGAATGGTTTGATACCGGAGCACGCACTTCTTCACATGAGGCCAAGGAAACGAAGAGTTGGATTAGTTTCAGAAAGACCTATAAGTCCTCGTACGGAAGAGGACAAATTAGCAGGAAAATTAGAAATCAAACGATATAAACAGACTAAAGAGAAACTTCGAAGACGTCGTGAGAAATTACTGCAGAAGGTAATGCTGGCTTCCGGGAAACTCAAACTCAGACCTGGACAGGCAAAATTACCCCTGTTAGACTTATCCGAGATAGACAGCGGGAGTGAATCGGATTCGGACGAGGCGGAGAAGTTGTGCTCTGGGTCGGCGAGCGTGCCTCCCGCTGTCCCCGCGCCCCCCGCCCCCGCCCCCGCCGCCGTGAGCGCGGAGACGTCGCGGCGCATCAAGGAGCAGTTCCGCAGCGCGATGGCGGGCGTCATGGTGCAGCACCTCAACCCGTACCGCCACTCGGACGCGCCGGCCGGCAGGATCACGTGCACCGCTGACTTTAAACACCTCGCCAGAAAGTTAACACACTTCGTAATGTTGAAAGAACTGAAACATTGCCGCTCCGTCGAGGAACTTGTCGTCACCGATTCTGTACGATCTAAAGCGAAGACTTTCGTCAAAAAATACATGGCCAAGTTCGGTCCGGTGTACAAACGGCCACCCGAGGAAGCAGACTAG
Protein
MSVEDSINDRNFLFQAEDSSSDEEEQLNSSLDSPLVMRHAKGKLVEGVNGIYEVIKEDAQNGLIPEHALLHMRPRKRRVGLVSERPISPRTEEDKLAGKLEIKRYKQTKEKLRRRREKLLQKVMLASGKLKLRPGQAKLPLLDLSEIDSGSESDSDEAEKLCSGSASVPPAVPAPPAPAPAAVSAETSRRIKEQFRSAMAGVMVQHLNPYRHSDAPAGRITCTADFKHLARKLTHFVMLKELKHCRSVEELVVTDSVRSKAKTFVKKYMAKFGPVYKRPPEEAD
Summary
Catalytic Activity
L-lysyl-[histone] + S-adenosyl-L-methionine = H(+) + N(6)-methyl-L-lysyl-[histone] + S-adenosyl-L-homocysteine
Uniprot
EC Number
2.1.1.43
EMBL
BABH01010259
KQ459598
KPI94599.1
KQ461183
KPJ07564.1
NEVH01019965
+ More
PNF22141.1 PNF22144.1 PNF22142.1 PNF22139.1 PNF22143.1 KK852415 KDR24367.1 KB632357 ERL93439.1 UFQT01004216 SSX35574.1 GEBQ01017729 JAT22248.1 UFQS01000436 UFQT01000436 SSX03918.1 SSX24283.1 APGK01040621 KB740984 ENN76193.1 KB630727 ERL83822.1 NNAY01004253 OXU18144.1 GEBQ01008113 JAT31864.1 KQ976394 KYM93355.1 ADTU01022133 ADTU01022134 KQ982268 KYQ58499.1 KQ980983 KYN10781.1 KQ981891 KYN33825.1 GL888137 EGI66662.1 KQ414612 KOC69170.1 KZ288349 PBC27388.1 GL762576 EFZ21071.1
PNF22141.1 PNF22144.1 PNF22142.1 PNF22139.1 PNF22143.1 KK852415 KDR24367.1 KB632357 ERL93439.1 UFQT01004216 SSX35574.1 GEBQ01017729 JAT22248.1 UFQS01000436 UFQT01000436 SSX03918.1 SSX24283.1 APGK01040621 KB740984 ENN76193.1 KB630727 ERL83822.1 NNAY01004253 OXU18144.1 GEBQ01008113 JAT31864.1 KQ976394 KYM93355.1 ADTU01022133 ADTU01022134 KQ982268 KYQ58499.1 KQ980983 KYN10781.1 KQ981891 KYN33825.1 GL888137 EGI66662.1 KQ414612 KOC69170.1 KZ288349 PBC27388.1 GL762576 EFZ21071.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF51045
SSF51045
Gene 3D
CDD
ProteinModelPortal
PDB
2A7O
E-value=3.89353e-21,
Score=248
Ontologies
GO
PANTHER
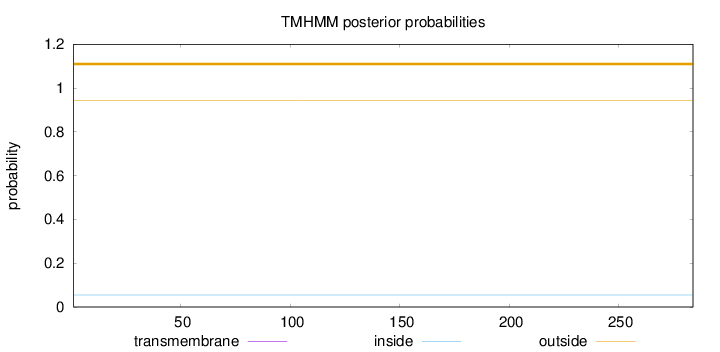
Topology
Subcellular location
Nucleus
Length:
284
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05519
outside
1 - 284
Population Genetic Test Statistics
Pi
23.559725
Theta
30.837677
Tajima's D
-1.629496
CLR
1.874091
CSRT
0.0427478626068697
Interpretation
Uncertain
