Gene
KWMTBOMO02233
Pre Gene Modal
BGIBMGA003111
Annotation
PREDICTED:_probable_serine_carboxypeptidase_CPVL_[Bombyx_mori]
Full name
Carboxypeptidase
+ More
Venom serine carboxypeptidase
Probable serine carboxypeptidase CPVL
Venom serine carboxypeptidase
Probable serine carboxypeptidase CPVL
Location in the cell
Extracellular Reliability : 1.725
Sequence
CDS
ATGTTGTTCATCGACAACCCTGTCGGCGCTGGCTTCAGCTTCACAGACAGTAACGATGGCTTCGTTAAAAATATGGATGAGTGCGCTTCGGATCTGTACACAGCTTTGAAACAATTCCTGATAATATTCCCCGAATTACGCACGGCACCACTGTACATTGCCGGTGAGTCGTACGCCGGGCGATACATTCCACCTTTTGCCATGAAATTGGCGGAGCAAGACCTCAACATTGGCACCTTTGAAGTCAATCTTCAGGGAATAATAATGGGAAATCCAGTGGTGGATCGTTACAGTGTTGCCAATTATACTTCCACGTTCCAGCAGTGGGGTCTAATAGACACGCAAGGAGCTATGTCAGTTAAATCTCTCCAAGATCAATACATTAAAGCCATATTGGACGGTGACGCGAGTGCTGCTTACGATTTACGCGACCAGTTGCTGTACAAACTGACTAAAATTACATTACAAAACCAATTTTTTAATGTACTAAAAAGCAATAAAGATCTTACGCTGTCTGAATTCGCGGCTTATCTACAAAAGAGTACAGTAAAAGAGGCACTCCACGTTGGAGACACAGATTTCTCCTTCATCAACACTACTGTAGACCTCGAGTTGAAGTCAGATTTCTTAACTGATATTGGAGGTACAGTCACAAAGTTATTAGAGCATTGCAAGGTATTAATATACTGCGGACAACTTGACTTGACCGCACCATGTGTACAGAGCGCCCAATATCGCAGACGCTATTGGCAGTGGAATAAAAAAGAACTTTTTCTTAGAGCGCCCAGAAGACCCTGGTTGCACAAGAATGTAGTAGCCGGGTACACCAAAAGTGGGGGTGGTTTTACAGAAGTTTTGGTGAAAGGAGCGGGTCATCTTGTACCCATAGACAAACCAGAGGAACTATACGAATTAATCAGCTGGTTTGTGTCAAGCCGAGAATTACTGCCATCTTCCGATTTCGACTCAGCTTCAGAATATGTTCCTGATTTAGATAAATCGTATTTTAGTTCTACGTCACCATCAGCAGAATCTGCATTGGGACCACAACTCGTAGCTAGATCAGCACACGGTCAACTCAGTCTTACAAAATCGGCAGAATCCAAAGATACCCCAATGTAA
Protein
MLFIDNPVGAGFSFTDSNDGFVKNMDECASDLYTALKQFLIIFPELRTAPLYIAGESYAGRYIPPFAMKLAEQDLNIGTFEVNLQGIIMGNPVVDRYSVANYTSTFQQWGLIDTQGAMSVKSLQDQYIKAILDGDASAAYDLRDQLLYKLTKITLQNQFFNVLKSNKDLTLSEFAAYLQKSTVKEALHVGDTDFSFINTTVDLELKSDFLTDIGGTVTKLLEHCKVLIYCGQLDLTAPCVQSAQYRRRYWQWNKKELFLRAPRRPWLHKNVVAGYTKSGGGFTEVLVKGAGHLVPIDKPEELYELISWFVSSRELLPSSDFDSASEYVPDLDKSYFSSTSPSAESALGPQLVARSAHGQLSLTKSAESKDTPM
Summary
Description
May be involved in the digestion of phagocytosed particles in the lysosome, participation in an inflammatory protease cascade, and trimming of peptides for antigen presentation.
Catalytic Activity
Release of a C-terminal amino acid with broad specificity.
Similarity
Belongs to the peptidase S10 family.
Keywords
Allergen
Carboxypeptidase
Complete proteome
Glycoprotein
Hydrolase
Protease
Reference proteome
Secreted
Signal
Alternative splicing
Zymogen
Feature
chain Carboxypeptidase
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9J0S4
A0A2H1V3Z0
A0A2A4K0D9
A0A0N1INC9
A0A194PNP2
A0A0N0P9M2
+ More
A0A1E1W9R5 A0A2P8YUZ9 A0A1W4X0Q1 A0A2R7WTA9 A0A2A4K0P1 A0A2H1V3X3 A0A146L8K2 K7IV07 H9J0S3 A0A348G5Z8 A0A195D9Z9 A0A195F7F6 R4WHX6 A0A2H1V444 A0A158ND03 A0A2A4JZQ9 A0A1Y1LKF5 A0A212EN12 A0A195B917 A0A195D1V5 E2BR37 F4X0L6 A0A0K8SFQ8 A0A224XJN6 A0A0A9W0I2 D7EHX7 A0A026WJK0 A0A151WYN0 A0A146L5J6 A0A0J7KEF9 A0A0K8SL40 B3SNT4 A0A170XJP7 E9IJU5 B7PI01 A0A0A9XV56 A0A1D2MBJ2 A0A2R7WYJ8 A0A220K8I1 A0A146LT64 A0A0A9XKA6 A0A146M3S2 C9WMM5 R4FLC5 A0A1V1G5G1 C1BPZ3 A0A2A3EHH5 H9J0S5 V5GQP5 A0A067QKM7 V5IG52 A0A1Y1KBB0 V5GN57 R4V3H3 U4U2S4 A0A1B0D870 A0A1B0D2X8 A0A2J7RDU7 A0A293N260 A0A1B6K7F1 A0A1V1FIY0 A0A0M8ZQT3 A0A0L7R680 A0A0A9XQP3 A0A2J7RMM3 A0A1B0CQ02 A0A0A9W9I9 A0A0K8SQH3 A0A146M8X8 A0A146L6S5 A0A146L0Y9 A0A2R7VXV6 A0A146M0Y4 A0A0A9YIS5 Q101N9 B7PC00 T1IBR4 K7IV06 E2AY00 A0A0A9XDE9 A0A0A9XJC2 B7ZN25 A0A212FM71 K7J463 R4FLQ2 A0A336M8H4 U5EN01 A0A0A9X8H2 Q9D3S9 Q869Q8 A0A067RMV2 B3SNT5 A0A1B6G2E3
A0A1E1W9R5 A0A2P8YUZ9 A0A1W4X0Q1 A0A2R7WTA9 A0A2A4K0P1 A0A2H1V3X3 A0A146L8K2 K7IV07 H9J0S3 A0A348G5Z8 A0A195D9Z9 A0A195F7F6 R4WHX6 A0A2H1V444 A0A158ND03 A0A2A4JZQ9 A0A1Y1LKF5 A0A212EN12 A0A195B917 A0A195D1V5 E2BR37 F4X0L6 A0A0K8SFQ8 A0A224XJN6 A0A0A9W0I2 D7EHX7 A0A026WJK0 A0A151WYN0 A0A146L5J6 A0A0J7KEF9 A0A0K8SL40 B3SNT4 A0A170XJP7 E9IJU5 B7PI01 A0A0A9XV56 A0A1D2MBJ2 A0A2R7WYJ8 A0A220K8I1 A0A146LT64 A0A0A9XKA6 A0A146M3S2 C9WMM5 R4FLC5 A0A1V1G5G1 C1BPZ3 A0A2A3EHH5 H9J0S5 V5GQP5 A0A067QKM7 V5IG52 A0A1Y1KBB0 V5GN57 R4V3H3 U4U2S4 A0A1B0D870 A0A1B0D2X8 A0A2J7RDU7 A0A293N260 A0A1B6K7F1 A0A1V1FIY0 A0A0M8ZQT3 A0A0L7R680 A0A0A9XQP3 A0A2J7RMM3 A0A1B0CQ02 A0A0A9W9I9 A0A0K8SQH3 A0A146M8X8 A0A146L6S5 A0A146L0Y9 A0A2R7VXV6 A0A146M0Y4 A0A0A9YIS5 Q101N9 B7PC00 T1IBR4 K7IV06 E2AY00 A0A0A9XDE9 A0A0A9XJC2 B7ZN25 A0A212FM71 K7J463 R4FLQ2 A0A336M8H4 U5EN01 A0A0A9X8H2 Q9D3S9 Q869Q8 A0A067RMV2 B3SNT5 A0A1B6G2E3
EC Number
3.4.16.-
3.4.16.5
3.4.16.5
Pubmed
EMBL
BABH01010273
BABH01010274
ODYU01000565
SOQ35547.1
NWSH01000324
PCG77476.1
+ More
KQ461183 KPJ07560.1 KQ459598 KPI94603.1 KQ459327 KPJ01438.1 GDQN01007312 JAT83742.1 PYGN01000345 PSN48057.1 KK855397 PTY22370.1 PCG77478.1 SOQ35545.1 GDHC01015187 JAQ03442.1 AAZX01001204 BABH01010267 BABH01010268 BABH01010269 BABH01010270 BABH01010271 FX985537 BBF97871.1 KQ981135 KYN09229.1 KQ981744 KYN36373.1 AK416960 BAN20175.1 SOQ35546.1 ADTU01012128 PCG77477.1 GEZM01053175 JAV74144.1 AGBW02013763 OWR42868.1 KQ976558 KYM80684.1 KQ977012 KYN06384.1 GL449898 EFN81843.1 GL888498 EGI60008.1 GBRD01013857 JAG51969.1 GFTR01006388 JAW10038.1 GBHO01045227 GBHO01045224 JAF98376.1 JAF98379.1 KQ971323 KYB28573.1 KK107213 QOIP01000007 EZA55299.1 RLU20576.1 KQ982650 KYQ52877.1 GDHC01016222 JAQ02407.1 LBMM01008690 KMQ88692.1 GBRD01011805 GBRD01011803 JAG54021.1 EF589781 ABU88379.2 GEMB01004337 JAR98936.1 GL763883 EFZ19168.1 ABJB010030144 ABJB010280903 DS716089 EEC06223.1 GBHO01022474 GBHO01022473 GBHO01022471 GBHO01003884 JAG21130.1 JAG21131.1 JAG21133.1 JAG39720.1 LJIJ01002009 ODM90366.1 KK855988 PTY24551.1 MF319693 ASJ26407.1 GDHC01008170 JAQ10459.1 GBHO01022472 JAG21132.1 GDHC01004208 JAQ14421.1 FJ765738 ACPB03001109 GAHY01001473 JAA76037.1 FX985403 BAX07416.1 BT076672 ACO11096.1 KZ288242 PBC31235.1 BABH01010276 BABH01010277 BABH01010278 BABH01010279 GANP01011728 JAB72740.1 KK853243 KDR09452.1 GANP01007292 JAB77176.1 GEZM01089787 JAV57440.1 GANP01012668 JAB71800.1 KC571839 AGM32338.1 KB631843 ERL86633.1 AJVK01027239 AJVK01023133 NEVH01005284 PNF39003.1 GFWV01022477 MAA47204.1 GECU01000337 JAT07370.1 FX985408 BAX07421.1 KQ435896 KOX69240.1 KQ414646 KOC66387.1 GBHO01022476 JAG21128.1 NEVH01002552 PNF42087.1 AJWK01022899 GBHO01039130 JAG04474.1 GBRD01010445 JAG55379.1 GDHC01002421 JAQ16208.1 GDHC01015334 JAQ03295.1 GDHC01016591 JAQ02038.1 KK854125 PTY11741.1 GDHC01005550 GDHC01000917 JAQ13079.1 JAQ17712.1 GBHO01028412 GBHO01014169 GBHO01004779 JAG15192.1 JAG29435.1 JAG38825.1 DQ104699 AAZ43093.2 ABJB010180836 ABJB010862162 ABJB010946688 ABJB011123252 DS680413 EEC04122.1 ACPB03018811 AAZX01001162 GL443740 EFN61703.1 GBHO01028515 GBHO01028514 JAG15089.1 JAG15090.1 GBHO01022777 JAG20827.1 BC144966 AAI44967.1 AGBW02007654 OWR54826.1 AAZX01003260 GAHY01002044 JAA75466.1 UFQT01000606 SSX25661.1 GANO01000788 JAB59083.1 GBHO01028516 GBHO01028513 GBRD01013050 JAG15088.1 JAG15091.1 JAG52776.1 AK017087 AC073297 AC079218 BC137839 AAFI02000012 KK852415 KDR24388.1 EF589782 ABU88380.2 GECZ01013161 JAS56608.1
KQ461183 KPJ07560.1 KQ459598 KPI94603.1 KQ459327 KPJ01438.1 GDQN01007312 JAT83742.1 PYGN01000345 PSN48057.1 KK855397 PTY22370.1 PCG77478.1 SOQ35545.1 GDHC01015187 JAQ03442.1 AAZX01001204 BABH01010267 BABH01010268 BABH01010269 BABH01010270 BABH01010271 FX985537 BBF97871.1 KQ981135 KYN09229.1 KQ981744 KYN36373.1 AK416960 BAN20175.1 SOQ35546.1 ADTU01012128 PCG77477.1 GEZM01053175 JAV74144.1 AGBW02013763 OWR42868.1 KQ976558 KYM80684.1 KQ977012 KYN06384.1 GL449898 EFN81843.1 GL888498 EGI60008.1 GBRD01013857 JAG51969.1 GFTR01006388 JAW10038.1 GBHO01045227 GBHO01045224 JAF98376.1 JAF98379.1 KQ971323 KYB28573.1 KK107213 QOIP01000007 EZA55299.1 RLU20576.1 KQ982650 KYQ52877.1 GDHC01016222 JAQ02407.1 LBMM01008690 KMQ88692.1 GBRD01011805 GBRD01011803 JAG54021.1 EF589781 ABU88379.2 GEMB01004337 JAR98936.1 GL763883 EFZ19168.1 ABJB010030144 ABJB010280903 DS716089 EEC06223.1 GBHO01022474 GBHO01022473 GBHO01022471 GBHO01003884 JAG21130.1 JAG21131.1 JAG21133.1 JAG39720.1 LJIJ01002009 ODM90366.1 KK855988 PTY24551.1 MF319693 ASJ26407.1 GDHC01008170 JAQ10459.1 GBHO01022472 JAG21132.1 GDHC01004208 JAQ14421.1 FJ765738 ACPB03001109 GAHY01001473 JAA76037.1 FX985403 BAX07416.1 BT076672 ACO11096.1 KZ288242 PBC31235.1 BABH01010276 BABH01010277 BABH01010278 BABH01010279 GANP01011728 JAB72740.1 KK853243 KDR09452.1 GANP01007292 JAB77176.1 GEZM01089787 JAV57440.1 GANP01012668 JAB71800.1 KC571839 AGM32338.1 KB631843 ERL86633.1 AJVK01027239 AJVK01023133 NEVH01005284 PNF39003.1 GFWV01022477 MAA47204.1 GECU01000337 JAT07370.1 FX985408 BAX07421.1 KQ435896 KOX69240.1 KQ414646 KOC66387.1 GBHO01022476 JAG21128.1 NEVH01002552 PNF42087.1 AJWK01022899 GBHO01039130 JAG04474.1 GBRD01010445 JAG55379.1 GDHC01002421 JAQ16208.1 GDHC01015334 JAQ03295.1 GDHC01016591 JAQ02038.1 KK854125 PTY11741.1 GDHC01005550 GDHC01000917 JAQ13079.1 JAQ17712.1 GBHO01028412 GBHO01014169 GBHO01004779 JAG15192.1 JAG29435.1 JAG38825.1 DQ104699 AAZ43093.2 ABJB010180836 ABJB010862162 ABJB010946688 ABJB011123252 DS680413 EEC04122.1 ACPB03018811 AAZX01001162 GL443740 EFN61703.1 GBHO01028515 GBHO01028514 JAG15089.1 JAG15090.1 GBHO01022777 JAG20827.1 BC144966 AAI44967.1 AGBW02007654 OWR54826.1 AAZX01003260 GAHY01002044 JAA75466.1 UFQT01000606 SSX25661.1 GANO01000788 JAB59083.1 GBHO01028516 GBHO01028513 GBRD01013050 JAG15088.1 JAG15091.1 JAG52776.1 AK017087 AC073297 AC079218 BC137839 AAFI02000012 KK852415 KDR24388.1 EF589782 ABU88380.2 GECZ01013161 JAS56608.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000245037
UP000192223
+ More
UP000002358 UP000078492 UP000078541 UP000005205 UP000007151 UP000078540 UP000078542 UP000008237 UP000007755 UP000007266 UP000053097 UP000279307 UP000075809 UP000036403 UP000001555 UP000094527 UP000005203 UP000015103 UP000242457 UP000027135 UP000030742 UP000092462 UP000235965 UP000053105 UP000053825 UP000092461 UP000000311 UP000000589 UP000002195
UP000002358 UP000078492 UP000078541 UP000005205 UP000007151 UP000078540 UP000078542 UP000008237 UP000007755 UP000007266 UP000053097 UP000279307 UP000075809 UP000036403 UP000001555 UP000094527 UP000005203 UP000015103 UP000242457 UP000027135 UP000030742 UP000092462 UP000235965 UP000053105 UP000053825 UP000092461 UP000000311 UP000000589 UP000002195
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9J0S4
A0A2H1V3Z0
A0A2A4K0D9
A0A0N1INC9
A0A194PNP2
A0A0N0P9M2
+ More
A0A1E1W9R5 A0A2P8YUZ9 A0A1W4X0Q1 A0A2R7WTA9 A0A2A4K0P1 A0A2H1V3X3 A0A146L8K2 K7IV07 H9J0S3 A0A348G5Z8 A0A195D9Z9 A0A195F7F6 R4WHX6 A0A2H1V444 A0A158ND03 A0A2A4JZQ9 A0A1Y1LKF5 A0A212EN12 A0A195B917 A0A195D1V5 E2BR37 F4X0L6 A0A0K8SFQ8 A0A224XJN6 A0A0A9W0I2 D7EHX7 A0A026WJK0 A0A151WYN0 A0A146L5J6 A0A0J7KEF9 A0A0K8SL40 B3SNT4 A0A170XJP7 E9IJU5 B7PI01 A0A0A9XV56 A0A1D2MBJ2 A0A2R7WYJ8 A0A220K8I1 A0A146LT64 A0A0A9XKA6 A0A146M3S2 C9WMM5 R4FLC5 A0A1V1G5G1 C1BPZ3 A0A2A3EHH5 H9J0S5 V5GQP5 A0A067QKM7 V5IG52 A0A1Y1KBB0 V5GN57 R4V3H3 U4U2S4 A0A1B0D870 A0A1B0D2X8 A0A2J7RDU7 A0A293N260 A0A1B6K7F1 A0A1V1FIY0 A0A0M8ZQT3 A0A0L7R680 A0A0A9XQP3 A0A2J7RMM3 A0A1B0CQ02 A0A0A9W9I9 A0A0K8SQH3 A0A146M8X8 A0A146L6S5 A0A146L0Y9 A0A2R7VXV6 A0A146M0Y4 A0A0A9YIS5 Q101N9 B7PC00 T1IBR4 K7IV06 E2AY00 A0A0A9XDE9 A0A0A9XJC2 B7ZN25 A0A212FM71 K7J463 R4FLQ2 A0A336M8H4 U5EN01 A0A0A9X8H2 Q9D3S9 Q869Q8 A0A067RMV2 B3SNT5 A0A1B6G2E3
A0A1E1W9R5 A0A2P8YUZ9 A0A1W4X0Q1 A0A2R7WTA9 A0A2A4K0P1 A0A2H1V3X3 A0A146L8K2 K7IV07 H9J0S3 A0A348G5Z8 A0A195D9Z9 A0A195F7F6 R4WHX6 A0A2H1V444 A0A158ND03 A0A2A4JZQ9 A0A1Y1LKF5 A0A212EN12 A0A195B917 A0A195D1V5 E2BR37 F4X0L6 A0A0K8SFQ8 A0A224XJN6 A0A0A9W0I2 D7EHX7 A0A026WJK0 A0A151WYN0 A0A146L5J6 A0A0J7KEF9 A0A0K8SL40 B3SNT4 A0A170XJP7 E9IJU5 B7PI01 A0A0A9XV56 A0A1D2MBJ2 A0A2R7WYJ8 A0A220K8I1 A0A146LT64 A0A0A9XKA6 A0A146M3S2 C9WMM5 R4FLC5 A0A1V1G5G1 C1BPZ3 A0A2A3EHH5 H9J0S5 V5GQP5 A0A067QKM7 V5IG52 A0A1Y1KBB0 V5GN57 R4V3H3 U4U2S4 A0A1B0D870 A0A1B0D2X8 A0A2J7RDU7 A0A293N260 A0A1B6K7F1 A0A1V1FIY0 A0A0M8ZQT3 A0A0L7R680 A0A0A9XQP3 A0A2J7RMM3 A0A1B0CQ02 A0A0A9W9I9 A0A0K8SQH3 A0A146M8X8 A0A146L6S5 A0A146L0Y9 A0A2R7VXV6 A0A146M0Y4 A0A0A9YIS5 Q101N9 B7PC00 T1IBR4 K7IV06 E2AY00 A0A0A9XDE9 A0A0A9XJC2 B7ZN25 A0A212FM71 K7J463 R4FLQ2 A0A336M8H4 U5EN01 A0A0A9X8H2 Q9D3S9 Q869Q8 A0A067RMV2 B3SNT5 A0A1B6G2E3
PDB
4MWT
E-value=2.20916e-17,
Score=218
Ontologies
GO
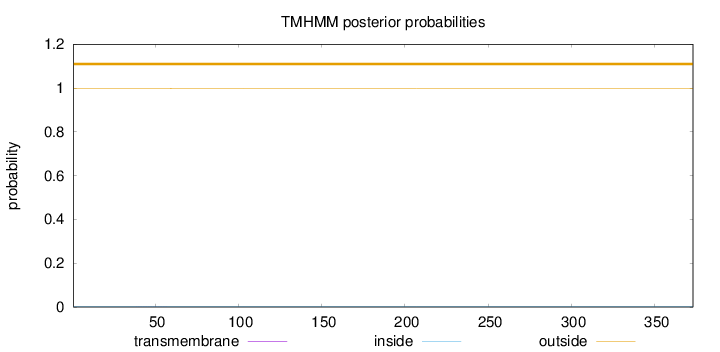
Topology
Subcellular location
Secreted
Length:
373
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00432999999999998
Exp number, first 60 AAs:
0.00346
Total prob of N-in:
0.00245
outside
1 - 373
Population Genetic Test Statistics
Pi
293.761057
Theta
179.476664
Tajima's D
2.47918
CLR
1.353339
CSRT
0.942952852357382
Interpretation
Uncertain
