Gene
KWMTBOMO02231 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003112
Annotation
PREDICTED:_transmembrane_9_superfamily_member_2_[Bombyx_mori]
Full name
Transmembrane 9 superfamily member
Location in the cell
PlasmaMembrane Reliability : 4.979
Sequence
CDS
ATGTCACTCCAGGCGTTTGTGATTTTTTCATTTCTGTTTTTAACACCTATTAAGGGTTTTTATTTACCTGGTTTAGCCCCCGTGAACTATTGCAAAACAACCGAAGATCCTGGAAAATCATGCAAGACAGAGATTCCTTTATATGTGAATAGGCTGAATACAGAAGAATCTATTATTCCATTTGAATACCACCACTTTGATTTCTGTACTACAAATGAGGACCAATCGCCTGTTGAAAATCTCGGTCAAGTAGTTTTTGGTGAGAGAATCCGTCCTAGTCCTTATAAACTAAAGTTTCTTGAGGATACAAAATGTAAAGCAGTATGCAAGAAGACATACAGAGGCTCCGATACAGATTCTACAAAAAAACTTAACTTGCTGAAGATGGGTATGACATTGTGCTACCAACATCACTGGATTTTGGATAATATGCCAGTGACCTGGTGTTACCTAGTCAATGAAGGTGGTAAAATGTACTGCAGTACTGGTTTTCCAATGGGTTGCCAAGTTCGTAAAGATAATATGGACACGTGTGCTTCGATAGTTACTATACCTGCAAAGGTTGGTGCCTATTACCTTGTTAACCATGTGGATCTGGAGATTACATACCACAGTGGTGGTGATGAAGATTGGGCTGTAAGCTTTGGTGAAAATGGAGGCCGCATTATATCTGCTAAGGTAAAGCCAGCCAGCATAAGACATGAAAACGAAAACAACTTAGACTGTACTGTTCGGAACAATCCTCAAGAAATACCAGCAACACTTGACAGTGATAAGACTTTGGACGTAATTTATACGTACAGTGTTACATTTATAAAGAACAACAACATTAAATGGTCATCGAGATGGGACTACATATTAGAATCAATGCCGCAGACCAATATCCAATGGTTCTCGATTTTAAACTCCTTGGTGATAGTCCTGTTCCTCAGCGGAATGGTTGCTATGATATTGCTAAGGACCCTCTATAAAGACATCGCTAGATACAACCAAATGGAATGTGGTGAAGATGCTCAGGAGGAATTCGGTTGGAAATTAGTTCACGGTGACGTCTTTCGACCTCCTCGTCGGGGCATGCTGCTGGCTGTCTTCTTAGGATCTGGATCTCAAGTGTTCGGCATGACACTTGTGACGCTGGCGTTCGCGTGCCTCGGGTTCCTGTCGCCGGCCAACCGCGGCGCGCTGATGACGTGCGCGCTCGTGGTGTGGGTGCTGCTGGGCGCCGCCGCCGGCTACGTCAGCGCGCGCGTCTACAAGAGCTTCGGCGGACGAAGATGGAAGAGCAATATCCTACTCACTTCCACTGTCTGTCCCGGCGTTGTTTTCTCTCTGTTCTTCGTGATGAACTTGGTGCTGTGGGGTAAGGACTCGTCGGCCGCGGTGCCGTTCTCGACGCTGGTGGCGCTGCTCGCCCTGTGGTTTGGAGTGTCTGTGCCGCTGACGTTCATCGGGGCCTACTTCGGCTTCAGAAAGAGGATCCTGGACCATCCAGTGCGCACCAACCAGATCCCGCGGCAGATCCCCGAGCAGTCCCTGTACACGCAGCCGATACCCGGCATCATCATGGGAGGAGTTTTGCCCTTCGGTTGCATCTTTATACAGCTGTTCTTCATACTCAACTCCCTGTGGTCTAGCCAGATGTACTATATGTTCGGCTTCCTGTTCCTGGTCTTCGTGATCCTGGTGATCACGTGTTCGGAGACGACGATCCTGCTGTGCTACTTCCACCTGTGCGCCGAGGACTACCACTGGTGGTGGCGCGCCTTCTTCAGCTCTGGCTCCACGGCCGGTTACCTGTTCATCTACTGCTGTCACTACTTCGTCACCAAGTTGAACATCGAGGACGCTGCCTCCACCTTCCTCTACTTCGGTTACACTCTGATCATGGTGTTCTTATTCTTCCTTCTGACCGGCACGATCGGCTTCATGGCCTGTTTCTGGTTCGTCAGGAAGATCTACAGCGTAGTGAAAGTAGATTAA
Protein
MSLQAFVIFSFLFLTPIKGFYLPGLAPVNYCKTTEDPGKSCKTEIPLYVNRLNTEESIIPFEYHHFDFCTTNEDQSPVENLGQVVFGERIRPSPYKLKFLEDTKCKAVCKKTYRGSDTDSTKKLNLLKMGMTLCYQHHWILDNMPVTWCYLVNEGGKMYCSTGFPMGCQVRKDNMDTCASIVTIPAKVGAYYLVNHVDLEITYHSGGDEDWAVSFGENGGRIISAKVKPASIRHENENNLDCTVRNNPQEIPATLDSDKTLDVIYTYSVTFIKNNNIKWSSRWDYILESMPQTNIQWFSILNSLVIVLFLSGMVAMILLRTLYKDIARYNQMECGEDAQEEFGWKLVHGDVFRPPRRGMLLAVFLGSGSQVFGMTLVTLAFACLGFLSPANRGALMTCALVVWVLLGAAAGYVSARVYKSFGGRRWKSNILLTSTVCPGVVFSLFFVMNLVLWGKDSSAAVPFSTLVALLALWFGVSVPLTFIGAYFGFRKRILDHPVRTNQIPRQIPEQSLYTQPIPGIIMGGVLPFGCIFIQLFFILNSLWSSQMYYMFGFLFLVFVILVITCSETTILLCYFHLCAEDYHWWWRAFFSSGSTAGYLFIYCCHYFVTKLNIEDAASTFLYFGYTLIMVFLFFLLTGTIGFMACFWFVRKIYSVVKVD
Summary
Similarity
Belongs to the nonaspanin (TM9SF) (TC 9.A.2) family.
Feature
chain Transmembrane 9 superfamily member
Uniprot
A0A2A4K0A4
A0A194PPE5
A0A084W8P4
A0A182Q392
A0A182K7K7
A0A182W8Z7
+ More
A0A2M4BGF5 A0A182XLU1 A0A182MRR2 A0A182V5K7 A0A182JEK9 A0A182YMR3 A0A182KYX0 A0A182HRG7 Q7PMH7 A0A182PG26 A0A182TUX9 A0A2M3YY11 A0A2M4A0V8 A0A182NAT6 W5JM57 A0A182F1P1 A0A182RR91 A0A0L7KXC5 U5EJZ0 A0A0M5J4A8 A0A1A9WLX6 A0A0L0C9X3 A0A1A9ZEX0 A0A1Q3FC35 A0A1Q3FE04 B4IFF1 B4Q3H1 A0A1W4UIJ5 A0A1A9YH29 A0A1A9VI47 A0A1B0BDQ1 A0A1Q3FE83 A0A1L8ECJ0 B0WBR6 A0A1Q3FE36 A0A1Q3FEN0 A0A1Q3FC72 A0A1L8ECK1 Q9VIK1 A0A1Q3FEM9 A0A1I8MRL3 A0A1I8PBG0 A0A1L8ECS8 A0A1L8ECN6 J9HZW5 A0A023EVR6 B3MK18 B4P6K8 A0A1L8DUQ3 A0A1L8DUP7 A0A1L8DUQ4 A0A1L8DUP9 T1E1E2 B4KJ02 B3NLY5 A0A1B6G6C7 A0A2J7QBN1 B4LV90 A0A2J7QBM2 B4MWB9 B4JAJ6 A0A3B0K3L4 V9IK88 Q29LW7 A0A154NYA6 A0A0L7R6W2 A0A2A3E9T9 A0A088A292 A0A1B0D268 T1PFR1 A0A067QQX4 A0A1L8EA40 K7IQ95 A0A151XDZ7 F4X4G2 A0A224XL54 A0A195BJ48 A0A158NJU0 A0A195F447 A0A146M6Y3 A0A0K8T2Y9 A0A0A9YMY9 A0A195DV60 E0VVM7 A0A026VWC5 A0A195CKC7 A0A3L8DNW4 W8BUQ4 E2BBS2 A0A1Y1LHN4 A0A2J7QBP1 R4WD57 V5GPY4 D6WKZ1
A0A2M4BGF5 A0A182XLU1 A0A182MRR2 A0A182V5K7 A0A182JEK9 A0A182YMR3 A0A182KYX0 A0A182HRG7 Q7PMH7 A0A182PG26 A0A182TUX9 A0A2M3YY11 A0A2M4A0V8 A0A182NAT6 W5JM57 A0A182F1P1 A0A182RR91 A0A0L7KXC5 U5EJZ0 A0A0M5J4A8 A0A1A9WLX6 A0A0L0C9X3 A0A1A9ZEX0 A0A1Q3FC35 A0A1Q3FE04 B4IFF1 B4Q3H1 A0A1W4UIJ5 A0A1A9YH29 A0A1A9VI47 A0A1B0BDQ1 A0A1Q3FE83 A0A1L8ECJ0 B0WBR6 A0A1Q3FE36 A0A1Q3FEN0 A0A1Q3FC72 A0A1L8ECK1 Q9VIK1 A0A1Q3FEM9 A0A1I8MRL3 A0A1I8PBG0 A0A1L8ECS8 A0A1L8ECN6 J9HZW5 A0A023EVR6 B3MK18 B4P6K8 A0A1L8DUQ3 A0A1L8DUP7 A0A1L8DUQ4 A0A1L8DUP9 T1E1E2 B4KJ02 B3NLY5 A0A1B6G6C7 A0A2J7QBN1 B4LV90 A0A2J7QBM2 B4MWB9 B4JAJ6 A0A3B0K3L4 V9IK88 Q29LW7 A0A154NYA6 A0A0L7R6W2 A0A2A3E9T9 A0A088A292 A0A1B0D268 T1PFR1 A0A067QQX4 A0A1L8EA40 K7IQ95 A0A151XDZ7 F4X4G2 A0A224XL54 A0A195BJ48 A0A158NJU0 A0A195F447 A0A146M6Y3 A0A0K8T2Y9 A0A0A9YMY9 A0A195DV60 E0VVM7 A0A026VWC5 A0A195CKC7 A0A3L8DNW4 W8BUQ4 E2BBS2 A0A1Y1LHN4 A0A2J7QBP1 R4WD57 V5GPY4 D6WKZ1
Pubmed
26354079
24438588
25244985
20966253
12364791
20920257
+ More
23761445 26227816 26108605 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17510324 24945155 26483478 17550304 24330624 18057021 15632085 24845553 20075255 21719571 21347285 26823975 25401762 20566863 24508170 30249741 24495485 20798317 28004739 23691247 18362917 19820115
23761445 26227816 26108605 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17510324 24945155 26483478 17550304 24330624 18057021 15632085 24845553 20075255 21719571 21347285 26823975 25401762 20566863 24508170 30249741 24495485 20798317 28004739 23691247 18362917 19820115
EMBL
NWSH01000324
PCG77479.1
KQ459598
KPI94604.1
ATLV01021497
KE525319
+ More
KFB46588.1 AXCN02000659 GGFJ01002900 MBW52041.1 AXCM01000135 APCN01001702 AAAB01008980 EAA13839.4 GGFM01000375 MBW21126.1 GGFK01001125 MBW34446.1 ADMH02001134 ETN63980.1 JTDY01004651 KOB67907.1 GANO01002016 JAB57855.1 CP012523 ALC40318.1 JRES01000714 KNC29010.1 GFDL01009928 JAV25117.1 GFDL01009254 JAV25791.1 CH480833 EDW46373.1 CM000361 CM002910 EDX05652.1 KMY91197.1 JXJN01012599 GFDL01009227 JAV25818.1 GFDG01002358 JAV16441.1 DS231881 EDS42687.1 GFDL01009238 JAV25807.1 GFDL01009041 JAV26004.1 GFDL01009869 JAV25176.1 GFDG01002348 JAV16451.1 AE014134 AY069665 AAF53917.1 AAL39810.1 GFDL01009040 JAV26005.1 GFDG01002357 JAV16442.1 GFDG01002332 JAV16467.1 CH478001 EJY58030.1 JXUM01023592 JXUM01023593 GAPW01000432 KQ560665 JAC13166.1 KXJ81377.1 CH902620 EDV31436.1 CM000158 EDW90960.1 GFDF01003925 JAV10159.1 GFDF01003922 JAV10162.1 GFDF01003924 JAV10160.1 GFDF01003923 JAV10161.1 GALA01001759 JAA93093.1 CH933807 EDW13515.2 KRG03796.1 CH954179 EDV54521.1 GECZ01022422 GECZ01011896 GECZ01002596 JAS47347.1 JAS57873.1 JAS67173.1 NEVH01016296 PNF25991.1 CH940649 EDW64350.2 PNF25990.1 CH963857 EDW75989.1 CH916368 EDW02782.1 OUUW01000004 SPP79621.1 JR049291 AEY60921.1 CH379060 EAL33928.2 KQ434775 KZC04064.1 KQ414645 KOC66627.1 KZ288311 PBC28488.1 AJVK01010496 AJVK01010497 KA646975 AFP61604.1 KK853129 KDR10991.1 GFDG01003159 JAV15640.1 KQ982254 KYQ58583.1 GL888644 EGI58672.1 GFTR01007665 JAW08761.1 KQ976465 KYM84359.1 ADTU01018377 KQ981820 KYN35355.1 GDHC01003430 JAQ15199.1 GBRD01005922 JAG59899.1 GBHO01010060 JAG33544.1 KQ980304 KYN16736.1 DS235812 EEB17433.1 KK107681 EZA48093.1 KQ977634 KYN01163.1 QOIP01000006 RLU22097.1 GAMC01001470 JAC05086.1 GL447151 EFN86848.1 GEZM01055102 JAV73144.1 PNF25989.1 AK417408 BAN20623.1 GALX01004804 JAB63662.1 KQ971343 EFA03539.1
KFB46588.1 AXCN02000659 GGFJ01002900 MBW52041.1 AXCM01000135 APCN01001702 AAAB01008980 EAA13839.4 GGFM01000375 MBW21126.1 GGFK01001125 MBW34446.1 ADMH02001134 ETN63980.1 JTDY01004651 KOB67907.1 GANO01002016 JAB57855.1 CP012523 ALC40318.1 JRES01000714 KNC29010.1 GFDL01009928 JAV25117.1 GFDL01009254 JAV25791.1 CH480833 EDW46373.1 CM000361 CM002910 EDX05652.1 KMY91197.1 JXJN01012599 GFDL01009227 JAV25818.1 GFDG01002358 JAV16441.1 DS231881 EDS42687.1 GFDL01009238 JAV25807.1 GFDL01009041 JAV26004.1 GFDL01009869 JAV25176.1 GFDG01002348 JAV16451.1 AE014134 AY069665 AAF53917.1 AAL39810.1 GFDL01009040 JAV26005.1 GFDG01002357 JAV16442.1 GFDG01002332 JAV16467.1 CH478001 EJY58030.1 JXUM01023592 JXUM01023593 GAPW01000432 KQ560665 JAC13166.1 KXJ81377.1 CH902620 EDV31436.1 CM000158 EDW90960.1 GFDF01003925 JAV10159.1 GFDF01003922 JAV10162.1 GFDF01003924 JAV10160.1 GFDF01003923 JAV10161.1 GALA01001759 JAA93093.1 CH933807 EDW13515.2 KRG03796.1 CH954179 EDV54521.1 GECZ01022422 GECZ01011896 GECZ01002596 JAS47347.1 JAS57873.1 JAS67173.1 NEVH01016296 PNF25991.1 CH940649 EDW64350.2 PNF25990.1 CH963857 EDW75989.1 CH916368 EDW02782.1 OUUW01000004 SPP79621.1 JR049291 AEY60921.1 CH379060 EAL33928.2 KQ434775 KZC04064.1 KQ414645 KOC66627.1 KZ288311 PBC28488.1 AJVK01010496 AJVK01010497 KA646975 AFP61604.1 KK853129 KDR10991.1 GFDG01003159 JAV15640.1 KQ982254 KYQ58583.1 GL888644 EGI58672.1 GFTR01007665 JAW08761.1 KQ976465 KYM84359.1 ADTU01018377 KQ981820 KYN35355.1 GDHC01003430 JAQ15199.1 GBRD01005922 JAG59899.1 GBHO01010060 JAG33544.1 KQ980304 KYN16736.1 DS235812 EEB17433.1 KK107681 EZA48093.1 KQ977634 KYN01163.1 QOIP01000006 RLU22097.1 GAMC01001470 JAC05086.1 GL447151 EFN86848.1 GEZM01055102 JAV73144.1 PNF25989.1 AK417408 BAN20623.1 GALX01004804 JAB63662.1 KQ971343 EFA03539.1
Proteomes
UP000218220
UP000053268
UP000030765
UP000075886
UP000075881
UP000075920
+ More
UP000076407 UP000075883 UP000075903 UP000075880 UP000076408 UP000075882 UP000075840 UP000007062 UP000075885 UP000075902 UP000075884 UP000000673 UP000069272 UP000075900 UP000037510 UP000092553 UP000091820 UP000037069 UP000092445 UP000001292 UP000000304 UP000192221 UP000092443 UP000078200 UP000092460 UP000002320 UP000000803 UP000095301 UP000095300 UP000008820 UP000069940 UP000249989 UP000007801 UP000002282 UP000009192 UP000008711 UP000235965 UP000008792 UP000007798 UP000001070 UP000268350 UP000001819 UP000076502 UP000053825 UP000242457 UP000005203 UP000092462 UP000027135 UP000002358 UP000075809 UP000007755 UP000078540 UP000005205 UP000078541 UP000078492 UP000009046 UP000053097 UP000078542 UP000279307 UP000008237 UP000007266
UP000076407 UP000075883 UP000075903 UP000075880 UP000076408 UP000075882 UP000075840 UP000007062 UP000075885 UP000075902 UP000075884 UP000000673 UP000069272 UP000075900 UP000037510 UP000092553 UP000091820 UP000037069 UP000092445 UP000001292 UP000000304 UP000192221 UP000092443 UP000078200 UP000092460 UP000002320 UP000000803 UP000095301 UP000095300 UP000008820 UP000069940 UP000249989 UP000007801 UP000002282 UP000009192 UP000008711 UP000235965 UP000008792 UP000007798 UP000001070 UP000268350 UP000001819 UP000076502 UP000053825 UP000242457 UP000005203 UP000092462 UP000027135 UP000002358 UP000075809 UP000007755 UP000078540 UP000005205 UP000078541 UP000078492 UP000009046 UP000053097 UP000078542 UP000279307 UP000008237 UP000007266
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K0A4
A0A194PPE5
A0A084W8P4
A0A182Q392
A0A182K7K7
A0A182W8Z7
+ More
A0A2M4BGF5 A0A182XLU1 A0A182MRR2 A0A182V5K7 A0A182JEK9 A0A182YMR3 A0A182KYX0 A0A182HRG7 Q7PMH7 A0A182PG26 A0A182TUX9 A0A2M3YY11 A0A2M4A0V8 A0A182NAT6 W5JM57 A0A182F1P1 A0A182RR91 A0A0L7KXC5 U5EJZ0 A0A0M5J4A8 A0A1A9WLX6 A0A0L0C9X3 A0A1A9ZEX0 A0A1Q3FC35 A0A1Q3FE04 B4IFF1 B4Q3H1 A0A1W4UIJ5 A0A1A9YH29 A0A1A9VI47 A0A1B0BDQ1 A0A1Q3FE83 A0A1L8ECJ0 B0WBR6 A0A1Q3FE36 A0A1Q3FEN0 A0A1Q3FC72 A0A1L8ECK1 Q9VIK1 A0A1Q3FEM9 A0A1I8MRL3 A0A1I8PBG0 A0A1L8ECS8 A0A1L8ECN6 J9HZW5 A0A023EVR6 B3MK18 B4P6K8 A0A1L8DUQ3 A0A1L8DUP7 A0A1L8DUQ4 A0A1L8DUP9 T1E1E2 B4KJ02 B3NLY5 A0A1B6G6C7 A0A2J7QBN1 B4LV90 A0A2J7QBM2 B4MWB9 B4JAJ6 A0A3B0K3L4 V9IK88 Q29LW7 A0A154NYA6 A0A0L7R6W2 A0A2A3E9T9 A0A088A292 A0A1B0D268 T1PFR1 A0A067QQX4 A0A1L8EA40 K7IQ95 A0A151XDZ7 F4X4G2 A0A224XL54 A0A195BJ48 A0A158NJU0 A0A195F447 A0A146M6Y3 A0A0K8T2Y9 A0A0A9YMY9 A0A195DV60 E0VVM7 A0A026VWC5 A0A195CKC7 A0A3L8DNW4 W8BUQ4 E2BBS2 A0A1Y1LHN4 A0A2J7QBP1 R4WD57 V5GPY4 D6WKZ1
A0A2M4BGF5 A0A182XLU1 A0A182MRR2 A0A182V5K7 A0A182JEK9 A0A182YMR3 A0A182KYX0 A0A182HRG7 Q7PMH7 A0A182PG26 A0A182TUX9 A0A2M3YY11 A0A2M4A0V8 A0A182NAT6 W5JM57 A0A182F1P1 A0A182RR91 A0A0L7KXC5 U5EJZ0 A0A0M5J4A8 A0A1A9WLX6 A0A0L0C9X3 A0A1A9ZEX0 A0A1Q3FC35 A0A1Q3FE04 B4IFF1 B4Q3H1 A0A1W4UIJ5 A0A1A9YH29 A0A1A9VI47 A0A1B0BDQ1 A0A1Q3FE83 A0A1L8ECJ0 B0WBR6 A0A1Q3FE36 A0A1Q3FEN0 A0A1Q3FC72 A0A1L8ECK1 Q9VIK1 A0A1Q3FEM9 A0A1I8MRL3 A0A1I8PBG0 A0A1L8ECS8 A0A1L8ECN6 J9HZW5 A0A023EVR6 B3MK18 B4P6K8 A0A1L8DUQ3 A0A1L8DUP7 A0A1L8DUQ4 A0A1L8DUP9 T1E1E2 B4KJ02 B3NLY5 A0A1B6G6C7 A0A2J7QBN1 B4LV90 A0A2J7QBM2 B4MWB9 B4JAJ6 A0A3B0K3L4 V9IK88 Q29LW7 A0A154NYA6 A0A0L7R6W2 A0A2A3E9T9 A0A088A292 A0A1B0D268 T1PFR1 A0A067QQX4 A0A1L8EA40 K7IQ95 A0A151XDZ7 F4X4G2 A0A224XL54 A0A195BJ48 A0A158NJU0 A0A195F447 A0A146M6Y3 A0A0K8T2Y9 A0A0A9YMY9 A0A195DV60 E0VVM7 A0A026VWC5 A0A195CKC7 A0A3L8DNW4 W8BUQ4 E2BBS2 A0A1Y1LHN4 A0A2J7QBP1 R4WD57 V5GPY4 D6WKZ1
Ontologies
GO
PANTHER
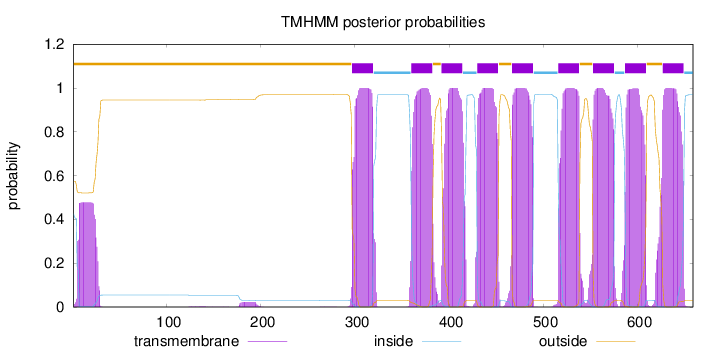
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.977502

Length:
659
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
216.40607
Exp number, first 60 AAs:
10.10853
Total prob of N-in:
0.42585
POSSIBLE N-term signal
sequence
outside
1 - 296
TMhelix
297 - 319
inside
320 - 359
TMhelix
360 - 382
outside
383 - 391
TMhelix
392 - 414
inside
415 - 429
TMhelix
430 - 452
outside
453 - 466
TMhelix
467 - 489
inside
490 - 515
TMhelix
516 - 538
outside
539 - 552
TMhelix
553 - 575
inside
576 - 586
TMhelix
587 - 609
outside
610 - 626
TMhelix
627 - 649
inside
650 - 659
Population Genetic Test Statistics
Pi
176.199301
Theta
159.383911
Tajima's D
0.051777
CLR
1.486992
CSRT
0.381730913454327
Interpretation
Uncertain
