Gene
KWMTBOMO02225
Pre Gene Modal
BGIBMGA003115
Annotation
PREDICTED:_peritrophin-48-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 4.68
Sequence
CDS
ATGTCTGCGCGCGCGCCAGCTTATCGCCCGGCTCTTGGCGTTCTCGTTGCGCTTGCTTTTGCCACGCGACTTGTCGGGAGTGATGACATTAACTGCACCAGAGACGGGCTTTATGCCGACTACGATAAAGAATGCAAAGAATACGTCCGTTGTTCCGGCGGAACAATCACTGGACGATATACGTGCAAATCAGGACGCAGTTTCAGTGAAGTTGCTGGTGCGTGTGTACCACAACAACGTCGACCGTGCGTCCGCCGCGTATGTACACCTGGAGATACCCTAGCCTATGCCATGCCCGGGACGGCTTGCAAGCATTATTACCGCTGCGAAAACGGAACTGCTGTTGATCGAGCATGTCCTTCCAGTACGTGGTTCGATCTCGACCGTCAAGCCTGCGCTCGAGGAGCCGGAACCTGTTATGAACCCGTTTGTGCCGGCCTCCCCGATGGTGAATATCCTGATTCATCGCACGAATGTCGTCGACTTTTACGTTGCGTCGGAGGAGAGTTGCGCGCTGTTGTATCGTGTACCAAGGATACCTGTGCTACTGCCTGTCCATCACCACGATCAGCCGCGGTGCCGTTACCGGCCGGCGATGCCGACTTCTGCTCCGACGAAGCTTGCTCGTCTTTATGCCAAAATCAACCTAATGGTGCTTACGCTGATCGCACTGCCGGATGTCGGGAATATTTCGTTTGCGAGACTCATCGAGTTATTCGTCGCGGTGTTTGCGAGCCCGGCATGCTATTCTCGAAAGCGGGTTGTGAACCAGCTACACGAGCGATATGCCCTCCGCCTGCGCTCAGTCCGTGTTTCAACCGTCCCGACGGCATGCATCGTGATTGGCGCTCTTGTTCATCGTGGTTCGATTGCAGAAGAGAGCGGGTAATTTCGAGAGGAACCTGCGCCACGGGGCTTGTCTTTGACGGGACAGGTTGCGTTCCGGCAACACAATTTCCATGCGATGGACCCGAACGATCTCAAGAGTGCGAAGGTCTTCCAAGTGGAACTTATCAAGATTTAGATTCAAACTGTACAAAATATTTCCATTGCGAGGGAGCTTCGCGCACTGTGCTCAGTTGTGGAGTCGGTCTTGTGTTTGACGGCGCTCGATGTTCACCTGCCGAACAATACTTGTGTCCGAGTCTAGAGAGAGACTCTTGCTACGGCAAAGCCGACGGTCGGTACCGCGCGGCGGACACTGGTTGTCGCGGGTACTACTCATGCGTGGCCGGTGAGAAATCTGTTTACGCGTGCCCATCTGGTAGTGTCTTTGATGATGAAGCCTGTGTGCCTGCGCAATCCGGCATTTGTCCCAGAGAAGATTACTCCTGTGACGGTTTATCAGACGGCTACCACCCAGAGATAGAATCCGGTTGCCATAGATATTTCTACTGCGAGGGCGGAGACCGACTAGCCACGCTGTCTTGCTTAGGAGGCAAAATATTCGACGGGCACGCGTGTGTGGAGACTCCGCACCACGAGTGCGGCGCGTTACCATCTAATTCAATCGAGCGCGGCGGTCCAGAATGTGAACGAGACGGATTCTTCGTGCAGGAGGGCACCGGCTGCAAACGTTACTACTTCTGTGTCAGCGGCAACAGAACGTACCTGACGTGTCCCGAACACAAAGTCTTCAACGGGCAGGTGTGCGTACCCAGTTCGCAGTACTCGTGTCCAGGGTGA
Protein
MSARAPAYRPALGVLVALAFATRLVGSDDINCTRDGLYADYDKECKEYVRCSGGTITGRYTCKSGRSFSEVAGACVPQQRRPCVRRVCTPGDTLAYAMPGTACKHYYRCENGTAVDRACPSSTWFDLDRQACARGAGTCYEPVCAGLPDGEYPDSSHECRRLLRCVGGELRAVVSCTKDTCATACPSPRSAAVPLPAGDADFCSDEACSSLCQNQPNGAYADRTAGCREYFVCETHRVIRRGVCEPGMLFSKAGCEPATRAICPPPALSPCFNRPDGMHRDWRSCSSWFDCRRERVISRGTCATGLVFDGTGCVPATQFPCDGPERSQECEGLPSGTYQDLDSNCTKYFHCEGASRTVLSCGVGLVFDGARCSPAEQYLCPSLERDSCYGKADGRYRAADTGCRGYYSCVAGEKSVYACPSGSVFDDEACVPAQSGICPREDYSCDGLSDGYHPEIESGCHRYFYCEGGDRLATLSCLGGKIFDGHACVETPHHECGALPSNSIERGGPECERDGFFVQEGTGCKRYYFCVSGNRTYLTCPEHKVFNGQVCVPSSQYSCPG
Summary
Uniprot
H9J0S8
A0A194PNP7
A0A0L7L5W9
A0A0N1IBK6
A0A212FDE0
A0A2H1VMN9
+ More
A0A2A4JZP1 A0A067QQV9 A0A2J7QBQ6 A0A2P8Y5M6 A0A2J7QBQ3 A0A1B6G024 Q17LW1 A0A1S4EY45 E0VVF1 A0A2S1ZS38 A0A182PDV4 A0A182V7B1 A0A182XNT0 A0A182LMQ8 A0A182JCS9 A0A1S4H0F1 A0A084VL29 A0A182I7D6 A0A182QL78 A0A182KBH9 A0A182Y0E1 A0A182W6B6 A0A182TQ41 A0A182MZ43 A0A182RI57 A0A182M3Z6 B0WB20 A0A182FN66 A0A2C9GQT4 A0A182T5A0 B7PMV9 A0A182U0T9 A0A182LNX9 A0A182WX55 A0A182PN30 A0A1S4HEA3 A0A182QSC0 A0A182RAM3 A0A182LVM6 A0A182JCC1 A0A182YLT0 A0A182MXN1 A0A182WEN4 A0A182M3K3 A0A084WGU1 A0A182WEN5 A0A182Q5E1 A0A1S4H868 A0A182FBG1 B4L019 A0A182M782 A0A182SK29 A0A2C9GR19 A0A182FDF6 A0A182XYN7 A0A182J762 A0A182WJF1 Q7PV23 W5JKC1
A0A2A4JZP1 A0A067QQV9 A0A2J7QBQ6 A0A2P8Y5M6 A0A2J7QBQ3 A0A1B6G024 Q17LW1 A0A1S4EY45 E0VVF1 A0A2S1ZS38 A0A182PDV4 A0A182V7B1 A0A182XNT0 A0A182LMQ8 A0A182JCS9 A0A1S4H0F1 A0A084VL29 A0A182I7D6 A0A182QL78 A0A182KBH9 A0A182Y0E1 A0A182W6B6 A0A182TQ41 A0A182MZ43 A0A182RI57 A0A182M3Z6 B0WB20 A0A182FN66 A0A2C9GQT4 A0A182T5A0 B7PMV9 A0A182U0T9 A0A182LNX9 A0A182WX55 A0A182PN30 A0A1S4HEA3 A0A182QSC0 A0A182RAM3 A0A182LVM6 A0A182JCC1 A0A182YLT0 A0A182MXN1 A0A182WEN4 A0A182M3K3 A0A084WGU1 A0A182WEN5 A0A182Q5E1 A0A1S4H868 A0A182FBG1 B4L019 A0A182M782 A0A182SK29 A0A2C9GR19 A0A182FDF6 A0A182XYN7 A0A182J762 A0A182WJF1 Q7PV23 W5JKC1
Pubmed
EMBL
BABH01010290
KQ459598
KPI94608.1
JTDY01002819
KOB70654.1
KQ461183
+ More
KPJ07554.1 AGBW02009066 OWR51761.1 ODYU01003391 SOQ42091.1 NWSH01000324 PCG77467.1 KK853129 KDR10966.1 NEVH01016296 PNF26016.1 PYGN01000900 PSN39541.1 PNF26017.1 GECZ01013987 JAS55782.1 CH477210 EAT47710.1 DS235811 EEB17357.1 MF942751 AWK28274.1 AAAB01008984 ATLV01014400 KE524970 KFB38673.1 APCN01003381 AXCN02000794 AXCM01006503 DS231876 EDS41978.1 APCN01001849 ABJB010031318 ABJB010247249 DS749294 EEC07931.1 AAAB01008986 AXCN02001054 AXCM01002955 ATLV01023716 KE525345 KFB49435.1 AXCN02000388 CH933809 EDW19054.2 AXCM01001523 EAA00608.5 ADMH02001280 ETN63224.1
KPJ07554.1 AGBW02009066 OWR51761.1 ODYU01003391 SOQ42091.1 NWSH01000324 PCG77467.1 KK853129 KDR10966.1 NEVH01016296 PNF26016.1 PYGN01000900 PSN39541.1 PNF26017.1 GECZ01013987 JAS55782.1 CH477210 EAT47710.1 DS235811 EEB17357.1 MF942751 AWK28274.1 AAAB01008984 ATLV01014400 KE524970 KFB38673.1 APCN01003381 AXCN02000794 AXCM01006503 DS231876 EDS41978.1 APCN01001849 ABJB010031318 ABJB010247249 DS749294 EEC07931.1 AAAB01008986 AXCN02001054 AXCM01002955 ATLV01023716 KE525345 KFB49435.1 AXCN02000388 CH933809 EDW19054.2 AXCM01001523 EAA00608.5 ADMH02001280 ETN63224.1
Proteomes
UP000005204
UP000053268
UP000037510
UP000053240
UP000007151
UP000218220
+ More
UP000027135 UP000235965 UP000245037 UP000008820 UP000009046 UP000075885 UP000075903 UP000076407 UP000075882 UP000075880 UP000030765 UP000075840 UP000075886 UP000075881 UP000076408 UP000075920 UP000075902 UP000075884 UP000075900 UP000075883 UP000002320 UP000069272 UP000075901 UP000001555 UP000009192 UP000007062 UP000000673
UP000027135 UP000235965 UP000245037 UP000008820 UP000009046 UP000075885 UP000075903 UP000076407 UP000075882 UP000075880 UP000030765 UP000075840 UP000075886 UP000075881 UP000076408 UP000075920 UP000075902 UP000075884 UP000075900 UP000075883 UP000002320 UP000069272 UP000075901 UP000001555 UP000009192 UP000007062 UP000000673
PRIDE
Pfam
PF01607 CBM_14
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
H9J0S8
A0A194PNP7
A0A0L7L5W9
A0A0N1IBK6
A0A212FDE0
A0A2H1VMN9
+ More
A0A2A4JZP1 A0A067QQV9 A0A2J7QBQ6 A0A2P8Y5M6 A0A2J7QBQ3 A0A1B6G024 Q17LW1 A0A1S4EY45 E0VVF1 A0A2S1ZS38 A0A182PDV4 A0A182V7B1 A0A182XNT0 A0A182LMQ8 A0A182JCS9 A0A1S4H0F1 A0A084VL29 A0A182I7D6 A0A182QL78 A0A182KBH9 A0A182Y0E1 A0A182W6B6 A0A182TQ41 A0A182MZ43 A0A182RI57 A0A182M3Z6 B0WB20 A0A182FN66 A0A2C9GQT4 A0A182T5A0 B7PMV9 A0A182U0T9 A0A182LNX9 A0A182WX55 A0A182PN30 A0A1S4HEA3 A0A182QSC0 A0A182RAM3 A0A182LVM6 A0A182JCC1 A0A182YLT0 A0A182MXN1 A0A182WEN4 A0A182M3K3 A0A084WGU1 A0A182WEN5 A0A182Q5E1 A0A1S4H868 A0A182FBG1 B4L019 A0A182M782 A0A182SK29 A0A2C9GR19 A0A182FDF6 A0A182XYN7 A0A182J762 A0A182WJF1 Q7PV23 W5JKC1
A0A2A4JZP1 A0A067QQV9 A0A2J7QBQ6 A0A2P8Y5M6 A0A2J7QBQ3 A0A1B6G024 Q17LW1 A0A1S4EY45 E0VVF1 A0A2S1ZS38 A0A182PDV4 A0A182V7B1 A0A182XNT0 A0A182LMQ8 A0A182JCS9 A0A1S4H0F1 A0A084VL29 A0A182I7D6 A0A182QL78 A0A182KBH9 A0A182Y0E1 A0A182W6B6 A0A182TQ41 A0A182MZ43 A0A182RI57 A0A182M3Z6 B0WB20 A0A182FN66 A0A2C9GQT4 A0A182T5A0 B7PMV9 A0A182U0T9 A0A182LNX9 A0A182WX55 A0A182PN30 A0A1S4HEA3 A0A182QSC0 A0A182RAM3 A0A182LVM6 A0A182JCC1 A0A182YLT0 A0A182MXN1 A0A182WEN4 A0A182M3K3 A0A084WGU1 A0A182WEN5 A0A182Q5E1 A0A1S4H868 A0A182FBG1 B4L019 A0A182M782 A0A182SK29 A0A2C9GR19 A0A182FDF6 A0A182XYN7 A0A182J762 A0A182WJF1 Q7PV23 W5JKC1
Ontologies
GO
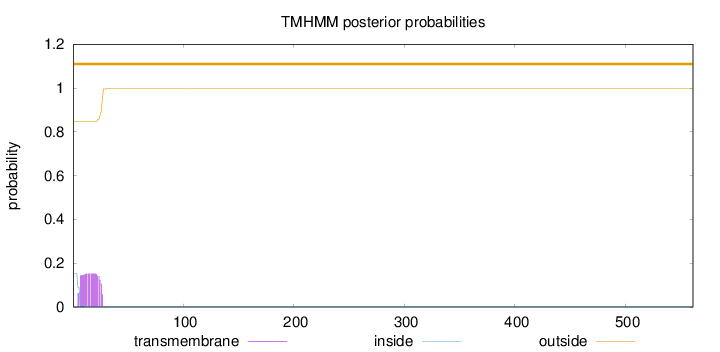
Topology
SignalP
Position: 1 - 27,
Likelihood: 0.988865
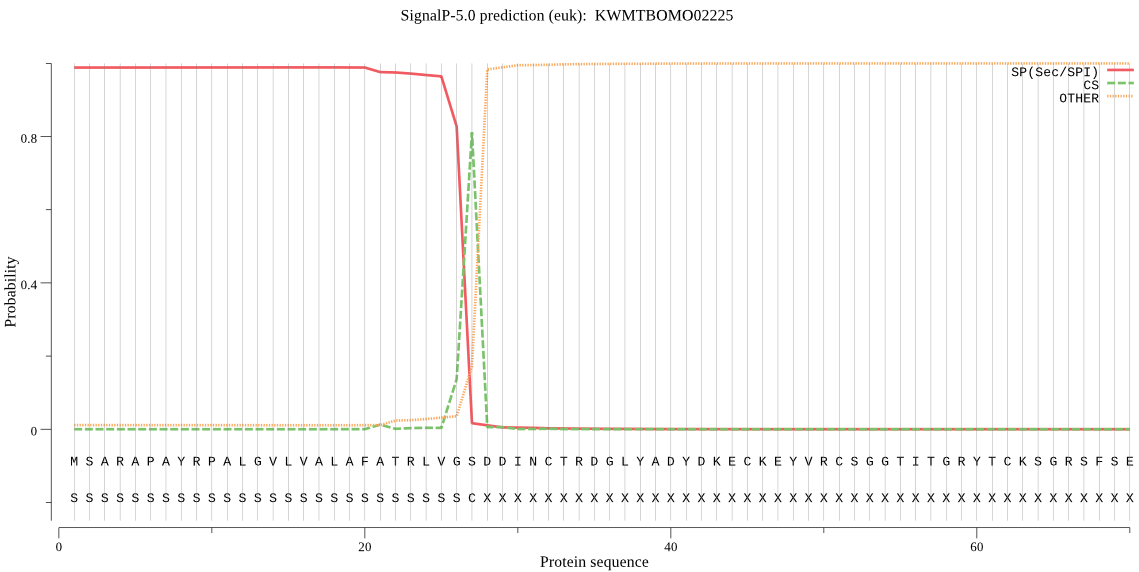
Length:
561
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.10781
Exp number, first 60 AAs:
3.10624
Total prob of N-in:
0.15228
outside
1 - 561
Population Genetic Test Statistics
Pi
174.744648
Theta
132.431217
Tajima's D
0.675097
CLR
24.664258
CSRT
0.566571671416429
Interpretation
Uncertain
