Gene
KWMTBOMO02223 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003116
Annotation
aspartate_aminotransferase_[Bombyx_mori]
Full name
Aspartate aminotransferase
Location in the cell
Cytoplasmic Reliability : 1.634 Mitochondrial Reliability : 1.806 Peroxisomal Reliability : 1.108
Sequence
CDS
ATGGCTCAAGTGTTAAAGAAATTAACTACCCAGGTGCTCAAGCCTAATAATGTAGACACGATCGTTGGTTGTACAGGCCTTAGAGCCAGCAGCACCTGGTGGAACAATGTTCAGATGGGTCCGCCAGATGTAATCTTAGGCATTACAGAAGCATACAAGAAGGACACACATCCTAAAAAAGTAAATCTTGGGGTTGGAGCATATAGAGACGATGAGGGAAAACCATTTGTTTTGCCTTCTGTCAGAAAGGCAGAAGAAATTCTTCACAGTAGAGGACTCAACCATGAGTATGCTCCCATTAGTGGTGAGGCCACATATACTGATGCTGTAGCTAAACTTGCTTTTGGAGAAGACAGTCCTGTCATCAAAAATAGGAGCAACTGTACTGTACAGACATTGTCCGGCACTGGCGCACTCCGACTCGGACTCGAGTTCATAACGAAACACTACGCTAAGGCAAAGGAGATCTGGCTCCCGACGCCTACTTGGGGAAACCACCCGCAAATCTGCAACACCCTCAACTTGCCGCACAAGAAATACCGTTACTTCGATCCCAAGACCAATGGTTTCGATTTGCAGGGAGCACTCGAGGATATATCAAAAATTCCCGAAGGTTCCATTATTCTGTTGCACGCGTGCGCACACAACCCCACCGGTGTTGACCCCAAGCCCAGCGACTGGGAACAACTTTCCAAGGTGATCAAAGAAAGGAAATTATTCCCGTTCTTCGACATGGCGTATCAAGGTTTCGCCACCGGTGATGTCGACAATGACGCTTTCGCAGTCCGTCTCTTCGTCAAGGAAGGACACCAAGTCATGTTGGCGCAGAGTTTCGCTAAGAACATGGGTCTGTACGGAGAGCGCGCGGGCGCCCTGACCTTCCTGTGCGGCGACGAGGCGACGGCCGCCAAGGTGATGTCGCAGGTCAAGATCATGGTCCGCGTCATGTACTCCAACCCTCCGCTGTACGGCGCCCGCCTCGTGCAGGAGATACTCACCAACGCCGAACTCAAAAAGCAATGGCTCGGAGACGTAAAGCAGATGGCGGACCGGATCATCACGATGCGGAGCCAGCTCCGCGCCGGCATCGAGGGCGCCGGCAACCCGCACCCCTGGCAACACATCACCGACCAGATCGGCATGTTCTGCTTCACCGGACTCAAGCCCGAGCAGGTCGAGCGTCTGACGAAAGAGTTCCACGTGTACCTGACGAAAGACGGACGCATTTCCGTCGCCGGCATCTCCTCGCAAAACGTGAACTACATCGCCGAGGCCATCCACAAGGTCACCTCATAA
Protein
MAQVLKKLTTQVLKPNNVDTIVGCTGLRASSTWWNNVQMGPPDVILGITEAYKKDTHPKKVNLGVGAYRDDEGKPFVLPSVRKAEEILHSRGLNHEYAPISGEATYTDAVAKLAFGEDSPVIKNRSNCTVQTLSGTGALRLGLEFITKHYAKAKEIWLPTPTWGNHPQICNTLNLPHKKYRYFDPKTNGFDLQGALEDISKIPEGSIILLHACAHNPTGVDPKPSDWEQLSKVIKERKLFPFFDMAYQGFATGDVDNDAFAVRLFVKEGHQVMLAQSFAKNMGLYGERAGALTFLCGDEATAAKVMSQVKIMVRVMYSNPPLYGARLVQEILTNAELKKQWLGDVKQMADRIITMRSQLRAGIEGAGNPHPWQHITDQIGMFCFTGLKPEQVERLTKEFHVYLTKDGRISVAGISSQNVNYIAEAIHKVTS
Summary
Catalytic Activity
2-oxoglutarate + L-aspartate = L-glutamate + oxaloacetate
Subunit
Homodimer.
Miscellaneous
In eukaryotes there are cytoplasmic, mitochondrial and chloroplastic isozymes.
Uniprot
Q1HQC7
A0A0N1I6L0
S4NIK5
A0A194PTZ8
I4DMJ4
A0A212FDH2
+ More
A0A0L8HF62 A0A346QRK7 A0A023FW96 T1D424 A0A023ES24 Q5K6H3 A0A1Z5KWT1 A0A023GM60 A0A023EUC0 G3MND0 A0A023FMK0 A0A293MXN9 A0A131XI81 L7M8A4 B0WHA4 A0A131Z1G6 A0A1Q3FI36 A0A1Q3FI61 A0A1E1XB04 A0A068CE32 A0A224YHY0 A0A182NBG9 A0A0P4WGM6 T1ELV3 A0A2R5LN70 V4ABY1 A0A182XN34 A0A182VTZ7 A0A2T7NHP2 A0A182PFJ4 A0A182VLB5 A0A182LM90 Q7PM06 A0A182HI77 A0A182U304 A0A084VXF8 R7VEL1 A0A182R3W1 A0A0U2USW7 A0A067QQX8 A0A182M173 A0A182QUS5 A0A182YFU6 A0A1B6LDY2 A0A0P4VSL5 R4G4U0 A0A210PZB0 A0A0B7AFG9 A0A0P5G423 A0A0P5K485 A0A1S3JTA5 A0A0N8AHJ2 A0A0P5GJ33 S5NFZ5 A0A2M4A4I2 A0A0P5MFN2 A0A0P5NA96 A0A182F252 K1R2Q9 W5J6A8 A0A0P6B3E8 A0A2M4A4A4 A0A232EMJ7 A0A0P5S2Y0 A0A0P4WPP3 A0A0P4X5Y2 A0A2J7QBK4 E2AHR4 A0A2M4BNR8 T1DND3 A0A182K5J8 A0A0P5SY60 A0A2M3Z217 A0A1B6IW93 A0A0P5HP03 A0A2C9K2S4 A0A023F820 A0A2M4BNN4 A0A182IP37 A0A170YUJ9 T1JUT6 A0A026VYP4 A0A3L8D4A3 A0A0P5MNK1 A0A0N8DE17 Q7ZWF5 A0A0P5G3V1 A0A0B7AET5 A0A0P5FYQ1 A0A1B6D614 A0A3B3CYX3 J3JT64 A0A0A9W0U8 F1QCD4
A0A0L8HF62 A0A346QRK7 A0A023FW96 T1D424 A0A023ES24 Q5K6H3 A0A1Z5KWT1 A0A023GM60 A0A023EUC0 G3MND0 A0A023FMK0 A0A293MXN9 A0A131XI81 L7M8A4 B0WHA4 A0A131Z1G6 A0A1Q3FI36 A0A1Q3FI61 A0A1E1XB04 A0A068CE32 A0A224YHY0 A0A182NBG9 A0A0P4WGM6 T1ELV3 A0A2R5LN70 V4ABY1 A0A182XN34 A0A182VTZ7 A0A2T7NHP2 A0A182PFJ4 A0A182VLB5 A0A182LM90 Q7PM06 A0A182HI77 A0A182U304 A0A084VXF8 R7VEL1 A0A182R3W1 A0A0U2USW7 A0A067QQX8 A0A182M173 A0A182QUS5 A0A182YFU6 A0A1B6LDY2 A0A0P4VSL5 R4G4U0 A0A210PZB0 A0A0B7AFG9 A0A0P5G423 A0A0P5K485 A0A1S3JTA5 A0A0N8AHJ2 A0A0P5GJ33 S5NFZ5 A0A2M4A4I2 A0A0P5MFN2 A0A0P5NA96 A0A182F252 K1R2Q9 W5J6A8 A0A0P6B3E8 A0A2M4A4A4 A0A232EMJ7 A0A0P5S2Y0 A0A0P4WPP3 A0A0P4X5Y2 A0A2J7QBK4 E2AHR4 A0A2M4BNR8 T1DND3 A0A182K5J8 A0A0P5SY60 A0A2M3Z217 A0A1B6IW93 A0A0P5HP03 A0A2C9K2S4 A0A023F820 A0A2M4BNN4 A0A182IP37 A0A170YUJ9 T1JUT6 A0A026VYP4 A0A3L8D4A3 A0A0P5MNK1 A0A0N8DE17 Q7ZWF5 A0A0P5G3V1 A0A0B7AET5 A0A0P5FYQ1 A0A1B6D614 A0A3B3CYX3 J3JT64 A0A0A9W0U8 F1QCD4
EC Number
2.6.1.1
Pubmed
26354079
23622113
22651552
22118469
24330624
24945155
+ More
17510324 28528879 22216098 28049606 25576852 26830274 28503490 28797301 23254933 20966253 12364791 14747013 17210077 24438588 26621068 24845553 25244985 27129103 28812685 23791183 22992520 20920257 23761445 28648823 20798317 15562597 25474469 24508170 30249741 29451363 22516182 23537049 25401762 23594743
17510324 28528879 22216098 28049606 25576852 26830274 28503490 28797301 23254933 20966253 12364791 14747013 17210077 24438588 26621068 24845553 25244985 27129103 28812685 23791183 22992520 20920257 23761445 28648823 20798317 15562597 25474469 24508170 30249741 29451363 22516182 23537049 25401762 23594743
EMBL
DQ443125
ABF51214.1
KQ461183
KPJ07552.1
GAIX01014034
JAA78526.1
+ More
KQ459598 KPI94610.1 AK402512 BAM19134.1 AGBW02009066 OWR51763.1 KQ418413 KOF87430.1 MH220515 AXR98597.1 GBBL01001189 JAC26131.1 GALA01001128 JAA93724.1 GAPW01001568 JAC12030.1 AF395206 CH477240 AAQ02892.1 EAT46448.1 GFJQ02007394 JAV99575.1 GBBM01001180 JAC34238.1 GAPW01001569 JAC12029.1 JO843381 AEO34998.1 GBBK01002544 JAC21938.1 GFWV01019960 MAA44688.1 GEFH01001738 JAP66843.1 GACK01004764 JAA60270.1 DS231934 EDS27610.1 GEDV01003293 JAP85264.1 GFDL01007837 JAV27208.1 GFDL01007805 JAV27240.1 GFAC01002887 JAT96301.1 KF773747 AID18683.1 GFPF01006131 MAA17277.1 GDRN01048331 JAI66661.1 AMQM01002880 KB095905 ESO10123.1 GGLE01006854 MBY10980.1 KB202325 ESO90816.1 PZQS01000012 PVD20691.1 AAAB01008980 EAA14551.4 APCN01005643 ATLV01018053 KE525212 KFB42652.1 AMQN01004791 KB294357 ELU14716.1 KT754869 ALS04703.1 KK853129 KDR10996.1 AXCM01000469 AXCN02000619 GEBQ01018087 JAT21890.1 GDKW01001135 JAI55460.1 ACPB03008949 GAHY01000471 JAA77039.1 NEDP02005352 OWF41825.1 HACG01032633 CEK79498.1 GDIQ01268313 LRGB01000626 JAJ83411.1 KZS17493.1 GDIQ01197458 GDIQ01189856 GDIQ01143642 JAK61869.1 GDIP01146801 JAJ76601.1 GDIQ01241782 JAK09943.1 KF021982 AGR65730.1 GGFK01002319 MBW35640.1 GDIQ01180112 JAK71613.1 GDIQ01145146 JAL06580.1 JH818674 EKC37844.1 ADMH02002076 ETN59536.1 GDIP01034349 JAM69366.1 GGFK01002323 MBW35644.1 NNAY01003371 OXU19532.1 GDIQ01097648 JAL54078.1 GDIP01254375 JAI69026.1 GDIP01254076 JAI69325.1 NEVH01016296 PNF25963.1 GL439579 EFN66993.1 GGFJ01005554 MBW54695.1 GAMD01002985 JAA98605.1 GDIP01136397 JAL67317.1 GGFM01001809 MBW22560.1 GECU01016507 JAS91199.1 GDIQ01225309 GDIQ01225308 JAK26416.1 GBBI01001324 JAC17388.1 GGFJ01005555 MBW54696.1 GEMB01002944 JAS00258.1 CAEY01000784 KK107727 EZA47979.1 QOIP01000013 RLU15337.1 GDIQ01160825 JAK90900.1 GDIP01042911 JAM60804.1 BC049435 AAH49435.1 GDIQ01246849 JAK04876.1 HACG01032634 CEK79499.1 GDIQ01248607 JAK03118.1 GEDC01016176 JAS21122.1 APGK01018933 BT126420 KB740085 KB632303 AEE61384.1 ENN81578.1 ERL91839.1 GBHO01045104 GBRD01004284 JAF98499.1 JAG61537.1 BX957362 CU633476
KQ459598 KPI94610.1 AK402512 BAM19134.1 AGBW02009066 OWR51763.1 KQ418413 KOF87430.1 MH220515 AXR98597.1 GBBL01001189 JAC26131.1 GALA01001128 JAA93724.1 GAPW01001568 JAC12030.1 AF395206 CH477240 AAQ02892.1 EAT46448.1 GFJQ02007394 JAV99575.1 GBBM01001180 JAC34238.1 GAPW01001569 JAC12029.1 JO843381 AEO34998.1 GBBK01002544 JAC21938.1 GFWV01019960 MAA44688.1 GEFH01001738 JAP66843.1 GACK01004764 JAA60270.1 DS231934 EDS27610.1 GEDV01003293 JAP85264.1 GFDL01007837 JAV27208.1 GFDL01007805 JAV27240.1 GFAC01002887 JAT96301.1 KF773747 AID18683.1 GFPF01006131 MAA17277.1 GDRN01048331 JAI66661.1 AMQM01002880 KB095905 ESO10123.1 GGLE01006854 MBY10980.1 KB202325 ESO90816.1 PZQS01000012 PVD20691.1 AAAB01008980 EAA14551.4 APCN01005643 ATLV01018053 KE525212 KFB42652.1 AMQN01004791 KB294357 ELU14716.1 KT754869 ALS04703.1 KK853129 KDR10996.1 AXCM01000469 AXCN02000619 GEBQ01018087 JAT21890.1 GDKW01001135 JAI55460.1 ACPB03008949 GAHY01000471 JAA77039.1 NEDP02005352 OWF41825.1 HACG01032633 CEK79498.1 GDIQ01268313 LRGB01000626 JAJ83411.1 KZS17493.1 GDIQ01197458 GDIQ01189856 GDIQ01143642 JAK61869.1 GDIP01146801 JAJ76601.1 GDIQ01241782 JAK09943.1 KF021982 AGR65730.1 GGFK01002319 MBW35640.1 GDIQ01180112 JAK71613.1 GDIQ01145146 JAL06580.1 JH818674 EKC37844.1 ADMH02002076 ETN59536.1 GDIP01034349 JAM69366.1 GGFK01002323 MBW35644.1 NNAY01003371 OXU19532.1 GDIQ01097648 JAL54078.1 GDIP01254375 JAI69026.1 GDIP01254076 JAI69325.1 NEVH01016296 PNF25963.1 GL439579 EFN66993.1 GGFJ01005554 MBW54695.1 GAMD01002985 JAA98605.1 GDIP01136397 JAL67317.1 GGFM01001809 MBW22560.1 GECU01016507 JAS91199.1 GDIQ01225309 GDIQ01225308 JAK26416.1 GBBI01001324 JAC17388.1 GGFJ01005555 MBW54696.1 GEMB01002944 JAS00258.1 CAEY01000784 KK107727 EZA47979.1 QOIP01000013 RLU15337.1 GDIQ01160825 JAK90900.1 GDIP01042911 JAM60804.1 BC049435 AAH49435.1 GDIQ01246849 JAK04876.1 HACG01032634 CEK79499.1 GDIQ01248607 JAK03118.1 GEDC01016176 JAS21122.1 APGK01018933 BT126420 KB740085 KB632303 AEE61384.1 ENN81578.1 ERL91839.1 GBHO01045104 GBRD01004284 JAF98499.1 JAG61537.1 BX957362 CU633476
Proteomes
UP000053240
UP000053268
UP000007151
UP000053454
UP000008820
UP000002320
+ More
UP000075884 UP000015101 UP000030746 UP000076407 UP000075920 UP000245119 UP000075885 UP000075903 UP000075882 UP000007062 UP000075840 UP000075902 UP000030765 UP000014760 UP000075900 UP000027135 UP000075883 UP000075886 UP000076408 UP000015103 UP000242188 UP000076858 UP000085678 UP000069272 UP000005408 UP000000673 UP000215335 UP000235965 UP000000311 UP000075881 UP000076420 UP000075880 UP000015104 UP000053097 UP000279307 UP000261560 UP000019118 UP000030742 UP000000437
UP000075884 UP000015101 UP000030746 UP000076407 UP000075920 UP000245119 UP000075885 UP000075903 UP000075882 UP000007062 UP000075840 UP000075902 UP000030765 UP000014760 UP000075900 UP000027135 UP000075883 UP000075886 UP000076408 UP000015103 UP000242188 UP000076858 UP000085678 UP000069272 UP000005408 UP000000673 UP000215335 UP000235965 UP000000311 UP000075881 UP000076420 UP000075880 UP000015104 UP000053097 UP000279307 UP000261560 UP000019118 UP000030742 UP000000437
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
Q1HQC7
A0A0N1I6L0
S4NIK5
A0A194PTZ8
I4DMJ4
A0A212FDH2
+ More
A0A0L8HF62 A0A346QRK7 A0A023FW96 T1D424 A0A023ES24 Q5K6H3 A0A1Z5KWT1 A0A023GM60 A0A023EUC0 G3MND0 A0A023FMK0 A0A293MXN9 A0A131XI81 L7M8A4 B0WHA4 A0A131Z1G6 A0A1Q3FI36 A0A1Q3FI61 A0A1E1XB04 A0A068CE32 A0A224YHY0 A0A182NBG9 A0A0P4WGM6 T1ELV3 A0A2R5LN70 V4ABY1 A0A182XN34 A0A182VTZ7 A0A2T7NHP2 A0A182PFJ4 A0A182VLB5 A0A182LM90 Q7PM06 A0A182HI77 A0A182U304 A0A084VXF8 R7VEL1 A0A182R3W1 A0A0U2USW7 A0A067QQX8 A0A182M173 A0A182QUS5 A0A182YFU6 A0A1B6LDY2 A0A0P4VSL5 R4G4U0 A0A210PZB0 A0A0B7AFG9 A0A0P5G423 A0A0P5K485 A0A1S3JTA5 A0A0N8AHJ2 A0A0P5GJ33 S5NFZ5 A0A2M4A4I2 A0A0P5MFN2 A0A0P5NA96 A0A182F252 K1R2Q9 W5J6A8 A0A0P6B3E8 A0A2M4A4A4 A0A232EMJ7 A0A0P5S2Y0 A0A0P4WPP3 A0A0P4X5Y2 A0A2J7QBK4 E2AHR4 A0A2M4BNR8 T1DND3 A0A182K5J8 A0A0P5SY60 A0A2M3Z217 A0A1B6IW93 A0A0P5HP03 A0A2C9K2S4 A0A023F820 A0A2M4BNN4 A0A182IP37 A0A170YUJ9 T1JUT6 A0A026VYP4 A0A3L8D4A3 A0A0P5MNK1 A0A0N8DE17 Q7ZWF5 A0A0P5G3V1 A0A0B7AET5 A0A0P5FYQ1 A0A1B6D614 A0A3B3CYX3 J3JT64 A0A0A9W0U8 F1QCD4
A0A0L8HF62 A0A346QRK7 A0A023FW96 T1D424 A0A023ES24 Q5K6H3 A0A1Z5KWT1 A0A023GM60 A0A023EUC0 G3MND0 A0A023FMK0 A0A293MXN9 A0A131XI81 L7M8A4 B0WHA4 A0A131Z1G6 A0A1Q3FI36 A0A1Q3FI61 A0A1E1XB04 A0A068CE32 A0A224YHY0 A0A182NBG9 A0A0P4WGM6 T1ELV3 A0A2R5LN70 V4ABY1 A0A182XN34 A0A182VTZ7 A0A2T7NHP2 A0A182PFJ4 A0A182VLB5 A0A182LM90 Q7PM06 A0A182HI77 A0A182U304 A0A084VXF8 R7VEL1 A0A182R3W1 A0A0U2USW7 A0A067QQX8 A0A182M173 A0A182QUS5 A0A182YFU6 A0A1B6LDY2 A0A0P4VSL5 R4G4U0 A0A210PZB0 A0A0B7AFG9 A0A0P5G423 A0A0P5K485 A0A1S3JTA5 A0A0N8AHJ2 A0A0P5GJ33 S5NFZ5 A0A2M4A4I2 A0A0P5MFN2 A0A0P5NA96 A0A182F252 K1R2Q9 W5J6A8 A0A0P6B3E8 A0A2M4A4A4 A0A232EMJ7 A0A0P5S2Y0 A0A0P4WPP3 A0A0P4X5Y2 A0A2J7QBK4 E2AHR4 A0A2M4BNR8 T1DND3 A0A182K5J8 A0A0P5SY60 A0A2M3Z217 A0A1B6IW93 A0A0P5HP03 A0A2C9K2S4 A0A023F820 A0A2M4BNN4 A0A182IP37 A0A170YUJ9 T1JUT6 A0A026VYP4 A0A3L8D4A3 A0A0P5MNK1 A0A0N8DE17 Q7ZWF5 A0A0P5G3V1 A0A0B7AET5 A0A0P5FYQ1 A0A1B6D614 A0A3B3CYX3 J3JT64 A0A0A9W0U8 F1QCD4
PDB
3PDB
E-value=1.16102e-154,
Score=1402
Ontologies
PATHWAY
00220
Arginine biosynthesis - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00350 Tyrosine metabolism - Bombyx mori (domestic silkworm)
00360 Phenylalanine metabolism - Bombyx mori (domestic silkworm)
00400 Phenylalanine, tyrosine and tryptophan biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00350 Tyrosine metabolism - Bombyx mori (domestic silkworm)
00360 Phenylalanine metabolism - Bombyx mori (domestic silkworm)
00400 Phenylalanine, tyrosine and tryptophan biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
PANTHER
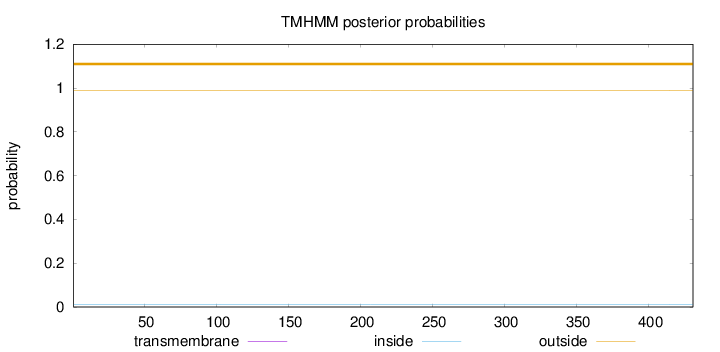
Topology
Length:
431
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00417
Exp number, first 60 AAs:
0.00206
Total prob of N-in:
0.01124
outside
1 - 431
Population Genetic Test Statistics
Pi
258.471175
Theta
175.543253
Tajima's D
0.822832
CLR
0.031577
CSRT
0.610469476526174
Interpretation
Uncertain
