Gene
KWMTBOMO02220
Pre Gene Modal
BGIBMGA003040
Annotation
PREDICTED:_uncharacterized_protein_LOC101740515_isoform_X2_[Bombyx_mori]
Transcription factor
Location in the cell
Mitochondrial Reliability : 1.942 Nuclear Reliability : 1.825
Sequence
CDS
ATGGTCAGAAAATACGAAAGAATGAGCGGCCGTCAGTCCTGGAATGAAGAAGATATGGCAAAGGCTGTCGCCGCTGTCGTCTCAGGTAAGATGGGCTACAAGTTGGCATCCAGGACCTACCATATACCGCGGTCTACTCTCCAGCGACGCGCCAGCAAGATACGGTATCAGCAACCAGACGAACCCAAGCCACTAATGGGGCACTACAGAAGGGTGTTCACTGATAGTCAAGAGAAAGACTTAGTTGGCTATATCAAAAGTATGGAAAAGTACTTTATGGGAGTCTCCCGAAGGGACATCAGAGAACTGGCATTTCAATACGCCGAAGACAACCATCTAAACCATCCCTTCGATGTAAACAGCCGAATGGCCGGAGAAGATTGGGTGCGAAACTTCCTGAAGCGCAATCCAGAACTCTTGGATAGATCCGACCACGAGAACATCCTAGAACCGGTTAATTTTGACCAATTCTATCATTTCATGTGTCAACTATCATGA
Protein
MVRKYERMSGRQSWNEEDMAKAVAAVVSGKMGYKLASRTYHIPRSTLQRRASKIRYQQPDEPKPLMGHYRRVFTDSQEKDLVGYIKSMEKYFMGVSRRDIRELAFQYAEDNHLNHPFDVNSRMAGEDWVRNFLKRNPELLDRSDHENILEPVNFDQFYHFMCQLS
Summary
Uniprot
H9J0K3
A0A2A4JB92
A0A212FHR3
A0A0L7KVG7
A0A194R3F7
A0A194PNQ2
+ More
S4PHB2 A0A1E1WAG7 A0A1B6L3T1 A0A1Y1MUL7 A0A1Y1K9Z4 A0A2H1VTA0 A0A1B6LLE7 A0A1Y1NJC0 A0A1Y1NGN4 A0A3Q0JIB2 A0A3L8DD48 J9LG75 J9JKJ5 A0A146KT86 A0A2H8TEI8 A0A1Y1JVK5 A0A1Y1NBL1 W4ZKV9 K7IN68 T1FFC3 T1FBR9 A0A232EV49 T1EWT4 T1FAC4 T1FGF6 T1EZV8 A0A212EU96 A0A1S3D1W0 T1F601 A0A1Y1LS13 T1END8 V4APS8 A0A1Y1LS50 A0A1B6JFQ6 A0A1Y1NBD5 K7JTE3 A0A1Y1LFM2 T1ICM1 J9KB99 T1FJV7 T1HKC3 T1F0Q2 A0A1Y1LR24 A0A2A4J0W1 A0A2P8XN66 A0A3L8DLV9 J9LBS3 A0A1Y1L660 A0A1W4XQ86 A0A0L7KY78 A0A1W4VE81 E9IWD6 A0A232F0H4 A0A0J7JY26 A0A0J7K472 A0A1S3DAF7 A0A232EDE8 V5GKW5 J9LWV0 A0A2A4J740 T1F5J1 T1EQM1 J9KKA5 A0A232FEU2 A0A1B6ME32 A0A0L7KY15 A0A2A4J792 K7J917 A0A2A4J6W1 A0A2G8LK40 B0X132 A0A1Y1M0X3 T1EJ72 A0A2A4J258 A0A2C9KFG9 A0A232EWE3 A0A0A9XB25 A0A2A4JD21 T1EJ73 A0A2A4J1M3 A0A2P8Y7N1 A0A1B6KS53 A0A2P8XAU1 A0A2A4JLU0 A0A2A4K2E5 A0A1B6D767 A0A026VT80 X1XGE9 A0A1B6CNF6 A0A1B6LHI0
S4PHB2 A0A1E1WAG7 A0A1B6L3T1 A0A1Y1MUL7 A0A1Y1K9Z4 A0A2H1VTA0 A0A1B6LLE7 A0A1Y1NJC0 A0A1Y1NGN4 A0A3Q0JIB2 A0A3L8DD48 J9LG75 J9JKJ5 A0A146KT86 A0A2H8TEI8 A0A1Y1JVK5 A0A1Y1NBL1 W4ZKV9 K7IN68 T1FFC3 T1FBR9 A0A232EV49 T1EWT4 T1FAC4 T1FGF6 T1EZV8 A0A212EU96 A0A1S3D1W0 T1F601 A0A1Y1LS13 T1END8 V4APS8 A0A1Y1LS50 A0A1B6JFQ6 A0A1Y1NBD5 K7JTE3 A0A1Y1LFM2 T1ICM1 J9KB99 T1FJV7 T1HKC3 T1F0Q2 A0A1Y1LR24 A0A2A4J0W1 A0A2P8XN66 A0A3L8DLV9 J9LBS3 A0A1Y1L660 A0A1W4XQ86 A0A0L7KY78 A0A1W4VE81 E9IWD6 A0A232F0H4 A0A0J7JY26 A0A0J7K472 A0A1S3DAF7 A0A232EDE8 V5GKW5 J9LWV0 A0A2A4J740 T1F5J1 T1EQM1 J9KKA5 A0A232FEU2 A0A1B6ME32 A0A0L7KY15 A0A2A4J792 K7J917 A0A2A4J6W1 A0A2G8LK40 B0X132 A0A1Y1M0X3 T1EJ72 A0A2A4J258 A0A2C9KFG9 A0A232EWE3 A0A0A9XB25 A0A2A4JD21 T1EJ73 A0A2A4J1M3 A0A2P8Y7N1 A0A1B6KS53 A0A2P8XAU1 A0A2A4JLU0 A0A2A4K2E5 A0A1B6D767 A0A026VT80 X1XGE9 A0A1B6CNF6 A0A1B6LHI0
Pubmed
EMBL
BABH01010298
NWSH01002118
PCG69119.1
AGBW02008477
OWR53256.1
JTDY01005368
+ More
KOB67076.1 KQ460779 KPJ12227.1 KQ459598 KPI94613.1 GAIX01005855 JAA86705.1 GDQN01007081 JAT83973.1 GEBQ01021595 JAT18382.1 GEZM01026324 JAV87047.1 GEZM01088155 JAV58299.1 ODYU01004314 SOQ44057.1 GEBQ01015395 JAT24582.1 GEZM01002998 JAV97000.1 GEZM01002997 JAV97001.1 QOIP01000010 RLU18063.1 ABLF02009840 ABLF02016204 ABLF02004627 ABLF02004628 ABLF02034930 GDHC01018928 JAP99700.1 GFXV01000711 MBW12516.1 GEZM01099403 GEZM01099402 JAV53344.1 GEZM01007307 JAV95253.1 AAGJ04133215 AAGJ04133216 AMQM01007068 KB097558 ESN94903.1 AMQM01006077 KB097269 ESN97849.1 NNAY01002052 OXU22234.1 AMQM01002059 KB097700 ESN91415.1 AMQM01005598 AMQM01005599 KB097026 ESN99962.1 AMQM01007412 KB097612 ESN93398.1 AMQM01002854 KB095905 ESO10016.1 AGBW02012433 OWR45073.1 AMQM01004377 KB096502 ESO04455.1 GEZM01048710 JAV76484.1 AMQM01000154 KB095811 ESO12383.1 KB201362 ESO96795.1 GEZM01048707 JAV76492.1 GECU01025373 GECU01009580 JAS82333.1 JAS98126.1 GEZM01007306 JAV95254.1 GEZM01058773 JAV71681.1 ACPB03009022 ABLF02022753 AMQM01008870 KB095865 ESO10484.1 ACPB03017843 AMQM01002986 KB095959 ESO09553.1 GEZM01050942 GEZM01050941 GEZM01050940 GEZM01050939 JAV75288.1 NWSH01004485 PCG65052.1 PYGN01001677 PSN33426.1 QOIP01000006 RLU21203.1 ABLF02026665 GEZM01067612 JAV67475.1 JTDY01004407 KOB68203.1 GL766495 EFZ15117.1 NNAY01001374 OXU24195.1 LBMM01021832 KMQ83002.1 LBMM01014670 KMQ85084.1 NNAY01006779 OXU16358.1 GALX01007438 JAB61028.1 ABLF02010765 ABLF02010766 NWSH01002782 PCG67546.1 AMQM01004277 ESO04218.1 AMQM01000642 KB096324 ESO06516.1 ABLF02006839 NNAY01000314 OXU29281.1 GEBQ01005828 JAT34149.1 JTDY01004613 KOB67946.1 NWSH01002570 PCG67955.1 AAZX01006942 NWSH01002933 PCG67263.1 PCG67264.1 MRZV01000053 PIK60582.1 DS232251 EDS38424.1 GEZM01047343 JAV76877.1 AMQM01007182 KB097579 ESN93974.1 NWSH01003844 PCG65768.1 NNAY01001872 OXU22675.1 GBHO01026440 JAG17164.1 NWSH01002054 PCG69293.1 AMQM01000552 ESO06091.1 PCG65769.1 PYGN01000838 PSN40164.1 GEBQ01025710 JAT14267.1 PYGN01006903 PSN29111.1 NWSH01001026 PCG73045.1 NWSH01000241 PCG78074.1 GEDC01015776 JAS21522.1 KK108004 EZA46988.1 ABLF02020045 GEDC01022355 JAS14943.1 GEBQ01016933 JAT23044.1
KOB67076.1 KQ460779 KPJ12227.1 KQ459598 KPI94613.1 GAIX01005855 JAA86705.1 GDQN01007081 JAT83973.1 GEBQ01021595 JAT18382.1 GEZM01026324 JAV87047.1 GEZM01088155 JAV58299.1 ODYU01004314 SOQ44057.1 GEBQ01015395 JAT24582.1 GEZM01002998 JAV97000.1 GEZM01002997 JAV97001.1 QOIP01000010 RLU18063.1 ABLF02009840 ABLF02016204 ABLF02004627 ABLF02004628 ABLF02034930 GDHC01018928 JAP99700.1 GFXV01000711 MBW12516.1 GEZM01099403 GEZM01099402 JAV53344.1 GEZM01007307 JAV95253.1 AAGJ04133215 AAGJ04133216 AMQM01007068 KB097558 ESN94903.1 AMQM01006077 KB097269 ESN97849.1 NNAY01002052 OXU22234.1 AMQM01002059 KB097700 ESN91415.1 AMQM01005598 AMQM01005599 KB097026 ESN99962.1 AMQM01007412 KB097612 ESN93398.1 AMQM01002854 KB095905 ESO10016.1 AGBW02012433 OWR45073.1 AMQM01004377 KB096502 ESO04455.1 GEZM01048710 JAV76484.1 AMQM01000154 KB095811 ESO12383.1 KB201362 ESO96795.1 GEZM01048707 JAV76492.1 GECU01025373 GECU01009580 JAS82333.1 JAS98126.1 GEZM01007306 JAV95254.1 GEZM01058773 JAV71681.1 ACPB03009022 ABLF02022753 AMQM01008870 KB095865 ESO10484.1 ACPB03017843 AMQM01002986 KB095959 ESO09553.1 GEZM01050942 GEZM01050941 GEZM01050940 GEZM01050939 JAV75288.1 NWSH01004485 PCG65052.1 PYGN01001677 PSN33426.1 QOIP01000006 RLU21203.1 ABLF02026665 GEZM01067612 JAV67475.1 JTDY01004407 KOB68203.1 GL766495 EFZ15117.1 NNAY01001374 OXU24195.1 LBMM01021832 KMQ83002.1 LBMM01014670 KMQ85084.1 NNAY01006779 OXU16358.1 GALX01007438 JAB61028.1 ABLF02010765 ABLF02010766 NWSH01002782 PCG67546.1 AMQM01004277 ESO04218.1 AMQM01000642 KB096324 ESO06516.1 ABLF02006839 NNAY01000314 OXU29281.1 GEBQ01005828 JAT34149.1 JTDY01004613 KOB67946.1 NWSH01002570 PCG67955.1 AAZX01006942 NWSH01002933 PCG67263.1 PCG67264.1 MRZV01000053 PIK60582.1 DS232251 EDS38424.1 GEZM01047343 JAV76877.1 AMQM01007182 KB097579 ESN93974.1 NWSH01003844 PCG65768.1 NNAY01001872 OXU22675.1 GBHO01026440 JAG17164.1 NWSH01002054 PCG69293.1 AMQM01000552 ESO06091.1 PCG65769.1 PYGN01000838 PSN40164.1 GEBQ01025710 JAT14267.1 PYGN01006903 PSN29111.1 NWSH01001026 PCG73045.1 NWSH01000241 PCG78074.1 GEDC01015776 JAS21522.1 KK108004 EZA46988.1 ABLF02020045 GEDC01022355 JAS14943.1 GEBQ01016933 JAT23044.1
Proteomes
PRIDE
Interpro
IPR007889
HTH_Psq
+ More
IPR009057 Homeobox-like_sf
IPR006600 HTH_CenpB_DNA-bd_dom
IPR004875 DDE_SF_endonuclease_dom
IPR011011 Znf_FYVE_PHD
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR011010 DNA_brk_join_enz
IPR021893 DUF3504
IPR013762 Integrase-like_cat_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR036880 Kunitz_BPTI_sf
IPR036388 WH-like_DNA-bd_sf
IPR007110 Ig-like_dom
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR009057 Homeobox-like_sf
IPR006600 HTH_CenpB_DNA-bd_dom
IPR004875 DDE_SF_endonuclease_dom
IPR011011 Znf_FYVE_PHD
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR011010 DNA_brk_join_enz
IPR021893 DUF3504
IPR013762 Integrase-like_cat_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR036880 Kunitz_BPTI_sf
IPR036388 WH-like_DNA-bd_sf
IPR007110 Ig-like_dom
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
SUPFAM
Gene 3D
ProteinModelPortal
H9J0K3
A0A2A4JB92
A0A212FHR3
A0A0L7KVG7
A0A194R3F7
A0A194PNQ2
+ More
S4PHB2 A0A1E1WAG7 A0A1B6L3T1 A0A1Y1MUL7 A0A1Y1K9Z4 A0A2H1VTA0 A0A1B6LLE7 A0A1Y1NJC0 A0A1Y1NGN4 A0A3Q0JIB2 A0A3L8DD48 J9LG75 J9JKJ5 A0A146KT86 A0A2H8TEI8 A0A1Y1JVK5 A0A1Y1NBL1 W4ZKV9 K7IN68 T1FFC3 T1FBR9 A0A232EV49 T1EWT4 T1FAC4 T1FGF6 T1EZV8 A0A212EU96 A0A1S3D1W0 T1F601 A0A1Y1LS13 T1END8 V4APS8 A0A1Y1LS50 A0A1B6JFQ6 A0A1Y1NBD5 K7JTE3 A0A1Y1LFM2 T1ICM1 J9KB99 T1FJV7 T1HKC3 T1F0Q2 A0A1Y1LR24 A0A2A4J0W1 A0A2P8XN66 A0A3L8DLV9 J9LBS3 A0A1Y1L660 A0A1W4XQ86 A0A0L7KY78 A0A1W4VE81 E9IWD6 A0A232F0H4 A0A0J7JY26 A0A0J7K472 A0A1S3DAF7 A0A232EDE8 V5GKW5 J9LWV0 A0A2A4J740 T1F5J1 T1EQM1 J9KKA5 A0A232FEU2 A0A1B6ME32 A0A0L7KY15 A0A2A4J792 K7J917 A0A2A4J6W1 A0A2G8LK40 B0X132 A0A1Y1M0X3 T1EJ72 A0A2A4J258 A0A2C9KFG9 A0A232EWE3 A0A0A9XB25 A0A2A4JD21 T1EJ73 A0A2A4J1M3 A0A2P8Y7N1 A0A1B6KS53 A0A2P8XAU1 A0A2A4JLU0 A0A2A4K2E5 A0A1B6D767 A0A026VT80 X1XGE9 A0A1B6CNF6 A0A1B6LHI0
S4PHB2 A0A1E1WAG7 A0A1B6L3T1 A0A1Y1MUL7 A0A1Y1K9Z4 A0A2H1VTA0 A0A1B6LLE7 A0A1Y1NJC0 A0A1Y1NGN4 A0A3Q0JIB2 A0A3L8DD48 J9LG75 J9JKJ5 A0A146KT86 A0A2H8TEI8 A0A1Y1JVK5 A0A1Y1NBL1 W4ZKV9 K7IN68 T1FFC3 T1FBR9 A0A232EV49 T1EWT4 T1FAC4 T1FGF6 T1EZV8 A0A212EU96 A0A1S3D1W0 T1F601 A0A1Y1LS13 T1END8 V4APS8 A0A1Y1LS50 A0A1B6JFQ6 A0A1Y1NBD5 K7JTE3 A0A1Y1LFM2 T1ICM1 J9KB99 T1FJV7 T1HKC3 T1F0Q2 A0A1Y1LR24 A0A2A4J0W1 A0A2P8XN66 A0A3L8DLV9 J9LBS3 A0A1Y1L660 A0A1W4XQ86 A0A0L7KY78 A0A1W4VE81 E9IWD6 A0A232F0H4 A0A0J7JY26 A0A0J7K472 A0A1S3DAF7 A0A232EDE8 V5GKW5 J9LWV0 A0A2A4J740 T1F5J1 T1EQM1 J9KKA5 A0A232FEU2 A0A1B6ME32 A0A0L7KY15 A0A2A4J792 K7J917 A0A2A4J6W1 A0A2G8LK40 B0X132 A0A1Y1M0X3 T1EJ72 A0A2A4J258 A0A2C9KFG9 A0A232EWE3 A0A0A9XB25 A0A2A4JD21 T1EJ73 A0A2A4J1M3 A0A2P8Y7N1 A0A1B6KS53 A0A2P8XAU1 A0A2A4JLU0 A0A2A4K2E5 A0A1B6D767 A0A026VT80 X1XGE9 A0A1B6CNF6 A0A1B6LHI0
Ontologies
GO
Topology
Subcellular location
Nucleus
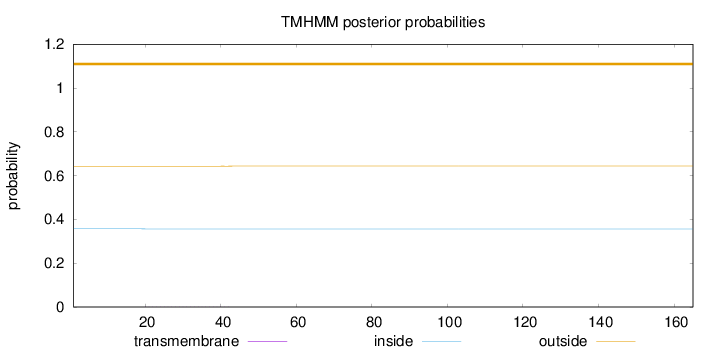
Length:
165
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01438
Exp number, first 60 AAs:
0.01438
Total prob of N-in:
0.35689
outside
1 - 165
Population Genetic Test Statistics
Pi
12.47138
Theta
11.897999
Tajima's D
-0.248502
CLR
1.437228
CSRT
0.307734613269337
Interpretation
Uncertain
