Pre Gene Modal
BGIBMGA002348
Annotation
elongation_factor_Ts_[Bombyx_mori]
Full name
Elongation factor Ts, mitochondrial
+ More
Elongation factor Ts
Elongation factor Ts
Location in the cell
Cytoplasmic Reliability : 2.089
Sequence
CDS
ATGATTTTCCAGCTGATTCGAAGAATTCATACAAGTCCAGCGTATAAAGCAGCTGAATCTTCGCTGCTTGCTAAACTCCGGAAAAAAACGGGCTACACAATCGCTAATTGCAAGAAAGCATTAGAAATGCATAATAATGATGCAGATAAGGCTGAGACATGGCTCCATGAACAAGCCCAAGCAATGGGATGGGCAAAGGCAACTAAATTAGCTGGCAGGACGGCCCTCCAGGGGTTGGTAGCTGTTAAGTTTGACAAGAACCATGGTGCTCTGGTGGAACTCAACTGTGAAACTGATTTTGTGGCCAAGAACGACAAGTTTCAGAAAATGATTGAAGATGCAGCTATAGCCTCATTCAAATTCGCTCATACTAAACTGCAAGCTAAAGGACCTATAACAAAAATGGAGCTAGACAGTGAACAGCTCGGGACATTACATGCAGAAGGTGGAAAAAAATTGTCTGAAATATTGGCACTCTTTATTGGCTCTGTCGGTGAAAATGCAGTGCTGAGGCGTGCCGAGTGCTGGAAAGCGAACAGCGAGGATGTCAAGATCGCAGGTTATACCCATCCAGCGCCTAGCAGTTCTAGTGATTATTCCTCTGGAAAATATGGGGCCCTGCTCGCTTACAAACAGCCAAACGACGTAGAAGACATCGGCCGGCAACTATGCCAGCACATCGTCGGCTGTGCACCCACAAAAATCGGTGACAAAGAAAAAGACAAACCAGCTAAGAACTCAGATGATGAAACATGTCTTATTTTCCAGGAATACTTGCTCGATCCATCCTATACGATTGAGGAAATCTTACAGAAACACAATGTTGAAATCATTGATTATGTTAGATTCTCTTGCGGAGAAGTACCTGAATCTTGTATGCCAGGAGTCGAAAAGCAACCTTTAGAAACAGTACAAACTCTGCAGTAA
Protein
MIFQLIRRIHTSPAYKAAESSLLAKLRKKTGYTIANCKKALEMHNNDADKAETWLHEQAQAMGWAKATKLAGRTALQGLVAVKFDKNHGALVELNCETDFVAKNDKFQKMIEDAAIASFKFAHTKLQAKGPITKMELDSEQLGTLHAEGGKKLSEILALFIGSVGENAVLRRAECWKANSEDVKIAGYTHPAPSSSSDYSSGKYGALLAYKQPNDVEDIGRQLCQHIVGCAPTKIGDKEKDKPAKNSDDETCLIFQEYLLDPSYTIEEILQKHNVEIIDYVRFSCGEVPESCMPGVEKQPLETVQTLQ
Summary
Description
Associates with the EF-Tu.GDP complex and induces the exchange of GDP to GTP. It remains bound to the aminoacyl-tRNA.EF-Tu.GTP complex up to the GTP hydrolysis stage on the ribosome.
Similarity
Belongs to the EF-Ts family.
Keywords
Complete proteome
Elongation factor
Mitochondrion
Protein biosynthesis
Reference proteome
Transit peptide
Feature
chain Elongation factor Ts, mitochondrial
Uniprot
Q1HQ90
A0A194PMF3
A0A194R3G3
A0A2A4JRG4
A0A2A4JRL6
A0A2H1W141
+ More
A0A212FHP8 A0A0L7LF35 S4PM30 Q17PI0 U5ES43 A0A182G5Y2 A0A067RJI5 K7IQU0 A0A232FBL8 A0A1Y1JYS9 B0WC25 B4Q7Z2 A0A1Q3FBV9 B4I5B7 A0A182MH27 A0A1W4UL41 A0A0L0C402 B4P8Q7 B3NMG3 Q9VJC7 A0A0C9R847 B4JC90 A0A182PAM8 A0A2M4BTT3 A0A2Y9D1U3 A0A2M4BTS4 A0A2M3Z7N6 D6WNZ5 A0A1W4X9X0 A0A182KH07 A0A2M4BSV0 A0A2M3Z7J7 A0A182QW97 A0A2M4CUH6 W5JT92 A0A1A9WZZ2 N6TZU7 A0A034WGZ4 A0A2M4ANH1 A0A2M4ANL1 A0A084VY74 T1DKY1 A0A2M4AP30 A0A182UAT7 A0A182JJN7 A0A1B0AJN3 A0A1S4H0G1 A0A182I5B4 A0A0K8VTW2 Q7PVN3 A0A0M3QTD0 A0A182VCT3 B4LSU6 A0A182Y9W9 T1PAS3 A0A1J1HPB4 A0A0A1WPW7 B4KEE0 A0A1A9UMG3 W8CBU2 A0A1I8M3T7 A0A1B0BRI3 A0A1A9Y773 A0A182WHB0 A0A1B0G3M0 B3MMH1 A0A182XKT1 A0A182NF87 A0A182FWG5 A0A151J8R1 E9J398 A0A182KU98 A0A1I8NR52 A0A026X5J8 A0A151I6P8 F4WU57 Q29LB7 B4GQA6 A0A0L7QVX8 A0A151X073 A0A151I028 E2C835 A0A2A3EIN6 A0A348G621 A0A158NLU2 A0A195FWQ9 A0A154P4V7 A0A3B0K922 E2AHM1 B4MYV9 A3EXL6 A0A088A1L8
A0A212FHP8 A0A0L7LF35 S4PM30 Q17PI0 U5ES43 A0A182G5Y2 A0A067RJI5 K7IQU0 A0A232FBL8 A0A1Y1JYS9 B0WC25 B4Q7Z2 A0A1Q3FBV9 B4I5B7 A0A182MH27 A0A1W4UL41 A0A0L0C402 B4P8Q7 B3NMG3 Q9VJC7 A0A0C9R847 B4JC90 A0A182PAM8 A0A2M4BTT3 A0A2Y9D1U3 A0A2M4BTS4 A0A2M3Z7N6 D6WNZ5 A0A1W4X9X0 A0A182KH07 A0A2M4BSV0 A0A2M3Z7J7 A0A182QW97 A0A2M4CUH6 W5JT92 A0A1A9WZZ2 N6TZU7 A0A034WGZ4 A0A2M4ANH1 A0A2M4ANL1 A0A084VY74 T1DKY1 A0A2M4AP30 A0A182UAT7 A0A182JJN7 A0A1B0AJN3 A0A1S4H0G1 A0A182I5B4 A0A0K8VTW2 Q7PVN3 A0A0M3QTD0 A0A182VCT3 B4LSU6 A0A182Y9W9 T1PAS3 A0A1J1HPB4 A0A0A1WPW7 B4KEE0 A0A1A9UMG3 W8CBU2 A0A1I8M3T7 A0A1B0BRI3 A0A1A9Y773 A0A182WHB0 A0A1B0G3M0 B3MMH1 A0A182XKT1 A0A182NF87 A0A182FWG5 A0A151J8R1 E9J398 A0A182KU98 A0A1I8NR52 A0A026X5J8 A0A151I6P8 F4WU57 Q29LB7 B4GQA6 A0A0L7QVX8 A0A151X073 A0A151I028 E2C835 A0A2A3EIN6 A0A348G621 A0A158NLU2 A0A195FWQ9 A0A154P4V7 A0A3B0K922 E2AHM1 B4MYV9 A3EXL6 A0A088A1L8
Pubmed
26354079
22118469
26227816
23622113
17510324
26483478
+ More
24845553 20075255 28648823 28004739 17994087 22936249 26108605 17550304 10731132 12537572 12537569 18362917 19820115 20920257 23761445 23537049 25348373 24438588 12364791 25244985 25830018 18057021 24495485 25315136 21282665 20966253 24508170 30249741 21719571 15632085 20798317 21347285
24845553 20075255 28648823 28004739 17994087 22936249 26108605 17550304 10731132 12537572 12537569 18362917 19820115 20920257 23761445 23537049 25348373 24438588 12364791 25244985 25830018 18057021 24495485 25315136 21282665 20966253 24508170 30249741 21719571 15632085 20798317 21347285
EMBL
DQ443162
ABF51251.1
KQ459598
KPI94616.1
KQ460779
KPJ12232.1
+ More
NWSH01000819 PCG74053.1 PCG74052.1 ODYU01005579 SOQ46562.1 AGBW02008477 OWR53259.1 JTDY01001388 KOB73994.1 GAIX01003760 JAA88800.1 CH477191 GANO01002531 JAB57340.1 JXUM01146978 KQ570274 KXJ68401.1 KK852432 KDR23982.1 NNAY01000461 OXU28221.1 GEZM01100259 JAV52940.1 DS231884 CM000361 CM002910 EDX05325.1 KMY90726.1 GFDL01010046 JAV24999.1 CH480822 EDW55573.1 AXCM01000590 JRES01000934 KNC27088.1 CM000158 EDW90165.1 CH954179 EDV54834.1 AE014134 AY051938 GBYB01007720 GBYB01012574 JAG77487.1 JAG82341.1 CH916368 EDW04123.1 GGFJ01007349 MBW56490.1 GGFJ01007262 MBW56403.1 GGFM01003773 MBW24524.1 KQ971343 EFA03187.2 GGFJ01006933 MBW56074.1 GGFM01003741 MBW24492.1 AXCN02000824 GGFL01004795 MBW68973.1 ADMH02000504 ETN66170.1 APGK01044884 APGK01044885 KB741028 KB631789 ENN74820.1 ERL86096.1 GAKP01005542 GAKP01005541 JAC53410.1 GGFK01009002 MBW42323.1 GGFK01009001 MBW42322.1 ATLV01018324 KE525231 KFB42918.1 GAMD01000711 JAB00880.1 GGFK01009208 MBW42529.1 AAAB01008984 APCN01003844 GDHF01010000 GDHF01001784 JAI42314.1 JAI50530.1 EAA14750.5 CP012523 ALC38669.1 CH940649 EDW64857.1 KA645842 AFP60471.1 CVRI01000015 CRK89896.1 GBXI01013829 GBXI01003789 JAD00463.1 JAD10503.1 CH933807 EDW12908.1 KRG03481.1 GAMC01004516 JAC02040.1 JXJN01019122 CCAG010019639 CH902620 EDV30917.1 KQ979548 KYN21071.1 GL768059 EFZ12704.1 KK107019 QOIP01000007 EZA62719.1 RLU20016.1 KQ978524 KYM93366.1 GL888349 EGI62284.1 CH379060 EAL34128.1 CH479187 EDW39778.1 KQ414722 KOC62699.1 KQ982635 KYQ53325.1 KQ976699 KYM77419.1 GL453538 EFN75888.1 KZ288250 PBC31069.1 FX985560 BBF97894.1 ADTU01019865 KQ981241 KYN44279.1 KQ434819 KZC06882.1 OUUW01000004 SPP80028.1 GL439566 EFN67055.1 CH963913 EDW77298.1 EF091934 ABN11926.1
NWSH01000819 PCG74053.1 PCG74052.1 ODYU01005579 SOQ46562.1 AGBW02008477 OWR53259.1 JTDY01001388 KOB73994.1 GAIX01003760 JAA88800.1 CH477191 GANO01002531 JAB57340.1 JXUM01146978 KQ570274 KXJ68401.1 KK852432 KDR23982.1 NNAY01000461 OXU28221.1 GEZM01100259 JAV52940.1 DS231884 CM000361 CM002910 EDX05325.1 KMY90726.1 GFDL01010046 JAV24999.1 CH480822 EDW55573.1 AXCM01000590 JRES01000934 KNC27088.1 CM000158 EDW90165.1 CH954179 EDV54834.1 AE014134 AY051938 GBYB01007720 GBYB01012574 JAG77487.1 JAG82341.1 CH916368 EDW04123.1 GGFJ01007349 MBW56490.1 GGFJ01007262 MBW56403.1 GGFM01003773 MBW24524.1 KQ971343 EFA03187.2 GGFJ01006933 MBW56074.1 GGFM01003741 MBW24492.1 AXCN02000824 GGFL01004795 MBW68973.1 ADMH02000504 ETN66170.1 APGK01044884 APGK01044885 KB741028 KB631789 ENN74820.1 ERL86096.1 GAKP01005542 GAKP01005541 JAC53410.1 GGFK01009002 MBW42323.1 GGFK01009001 MBW42322.1 ATLV01018324 KE525231 KFB42918.1 GAMD01000711 JAB00880.1 GGFK01009208 MBW42529.1 AAAB01008984 APCN01003844 GDHF01010000 GDHF01001784 JAI42314.1 JAI50530.1 EAA14750.5 CP012523 ALC38669.1 CH940649 EDW64857.1 KA645842 AFP60471.1 CVRI01000015 CRK89896.1 GBXI01013829 GBXI01003789 JAD00463.1 JAD10503.1 CH933807 EDW12908.1 KRG03481.1 GAMC01004516 JAC02040.1 JXJN01019122 CCAG010019639 CH902620 EDV30917.1 KQ979548 KYN21071.1 GL768059 EFZ12704.1 KK107019 QOIP01000007 EZA62719.1 RLU20016.1 KQ978524 KYM93366.1 GL888349 EGI62284.1 CH379060 EAL34128.1 CH479187 EDW39778.1 KQ414722 KOC62699.1 KQ982635 KYQ53325.1 KQ976699 KYM77419.1 GL453538 EFN75888.1 KZ288250 PBC31069.1 FX985560 BBF97894.1 ADTU01019865 KQ981241 KYN44279.1 KQ434819 KZC06882.1 OUUW01000004 SPP80028.1 GL439566 EFN67055.1 CH963913 EDW77298.1 EF091934 ABN11926.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
UP000008820
+ More
UP000069940 UP000249989 UP000027135 UP000002358 UP000215335 UP000002320 UP000000304 UP000001292 UP000075883 UP000192221 UP000037069 UP000002282 UP000008711 UP000000803 UP000001070 UP000075885 UP000075900 UP000007266 UP000192223 UP000075881 UP000075886 UP000000673 UP000091820 UP000019118 UP000030742 UP000030765 UP000075902 UP000075880 UP000092445 UP000075840 UP000007062 UP000092553 UP000075903 UP000008792 UP000076408 UP000183832 UP000009192 UP000078200 UP000095301 UP000092460 UP000092443 UP000075920 UP000092444 UP000007801 UP000076407 UP000075884 UP000069272 UP000078492 UP000075882 UP000095300 UP000053097 UP000279307 UP000078542 UP000007755 UP000001819 UP000008744 UP000053825 UP000075809 UP000078540 UP000008237 UP000242457 UP000005205 UP000078541 UP000076502 UP000268350 UP000000311 UP000007798 UP000005203
UP000069940 UP000249989 UP000027135 UP000002358 UP000215335 UP000002320 UP000000304 UP000001292 UP000075883 UP000192221 UP000037069 UP000002282 UP000008711 UP000000803 UP000001070 UP000075885 UP000075900 UP000007266 UP000192223 UP000075881 UP000075886 UP000000673 UP000091820 UP000019118 UP000030742 UP000030765 UP000075902 UP000075880 UP000092445 UP000075840 UP000007062 UP000092553 UP000075903 UP000008792 UP000076408 UP000183832 UP000009192 UP000078200 UP000095301 UP000092460 UP000092443 UP000075920 UP000092444 UP000007801 UP000076407 UP000075884 UP000069272 UP000078492 UP000075882 UP000095300 UP000053097 UP000279307 UP000078542 UP000007755 UP000001819 UP000008744 UP000053825 UP000075809 UP000078540 UP000008237 UP000242457 UP000005205 UP000078541 UP000076502 UP000268350 UP000000311 UP000007798 UP000005203
Interpro
Gene 3D
ProteinModelPortal
Q1HQ90
A0A194PMF3
A0A194R3G3
A0A2A4JRG4
A0A2A4JRL6
A0A2H1W141
+ More
A0A212FHP8 A0A0L7LF35 S4PM30 Q17PI0 U5ES43 A0A182G5Y2 A0A067RJI5 K7IQU0 A0A232FBL8 A0A1Y1JYS9 B0WC25 B4Q7Z2 A0A1Q3FBV9 B4I5B7 A0A182MH27 A0A1W4UL41 A0A0L0C402 B4P8Q7 B3NMG3 Q9VJC7 A0A0C9R847 B4JC90 A0A182PAM8 A0A2M4BTT3 A0A2Y9D1U3 A0A2M4BTS4 A0A2M3Z7N6 D6WNZ5 A0A1W4X9X0 A0A182KH07 A0A2M4BSV0 A0A2M3Z7J7 A0A182QW97 A0A2M4CUH6 W5JT92 A0A1A9WZZ2 N6TZU7 A0A034WGZ4 A0A2M4ANH1 A0A2M4ANL1 A0A084VY74 T1DKY1 A0A2M4AP30 A0A182UAT7 A0A182JJN7 A0A1B0AJN3 A0A1S4H0G1 A0A182I5B4 A0A0K8VTW2 Q7PVN3 A0A0M3QTD0 A0A182VCT3 B4LSU6 A0A182Y9W9 T1PAS3 A0A1J1HPB4 A0A0A1WPW7 B4KEE0 A0A1A9UMG3 W8CBU2 A0A1I8M3T7 A0A1B0BRI3 A0A1A9Y773 A0A182WHB0 A0A1B0G3M0 B3MMH1 A0A182XKT1 A0A182NF87 A0A182FWG5 A0A151J8R1 E9J398 A0A182KU98 A0A1I8NR52 A0A026X5J8 A0A151I6P8 F4WU57 Q29LB7 B4GQA6 A0A0L7QVX8 A0A151X073 A0A151I028 E2C835 A0A2A3EIN6 A0A348G621 A0A158NLU2 A0A195FWQ9 A0A154P4V7 A0A3B0K922 E2AHM1 B4MYV9 A3EXL6 A0A088A1L8
A0A212FHP8 A0A0L7LF35 S4PM30 Q17PI0 U5ES43 A0A182G5Y2 A0A067RJI5 K7IQU0 A0A232FBL8 A0A1Y1JYS9 B0WC25 B4Q7Z2 A0A1Q3FBV9 B4I5B7 A0A182MH27 A0A1W4UL41 A0A0L0C402 B4P8Q7 B3NMG3 Q9VJC7 A0A0C9R847 B4JC90 A0A182PAM8 A0A2M4BTT3 A0A2Y9D1U3 A0A2M4BTS4 A0A2M3Z7N6 D6WNZ5 A0A1W4X9X0 A0A182KH07 A0A2M4BSV0 A0A2M3Z7J7 A0A182QW97 A0A2M4CUH6 W5JT92 A0A1A9WZZ2 N6TZU7 A0A034WGZ4 A0A2M4ANH1 A0A2M4ANL1 A0A084VY74 T1DKY1 A0A2M4AP30 A0A182UAT7 A0A182JJN7 A0A1B0AJN3 A0A1S4H0G1 A0A182I5B4 A0A0K8VTW2 Q7PVN3 A0A0M3QTD0 A0A182VCT3 B4LSU6 A0A182Y9W9 T1PAS3 A0A1J1HPB4 A0A0A1WPW7 B4KEE0 A0A1A9UMG3 W8CBU2 A0A1I8M3T7 A0A1B0BRI3 A0A1A9Y773 A0A182WHB0 A0A1B0G3M0 B3MMH1 A0A182XKT1 A0A182NF87 A0A182FWG5 A0A151J8R1 E9J398 A0A182KU98 A0A1I8NR52 A0A026X5J8 A0A151I6P8 F4WU57 Q29LB7 B4GQA6 A0A0L7QVX8 A0A151X073 A0A151I028 E2C835 A0A2A3EIN6 A0A348G621 A0A158NLU2 A0A195FWQ9 A0A154P4V7 A0A3B0K922 E2AHM1 B4MYV9 A3EXL6 A0A088A1L8
PDB
1XB2
E-value=7.90232e-49,
Score=488
Ontologies
GO
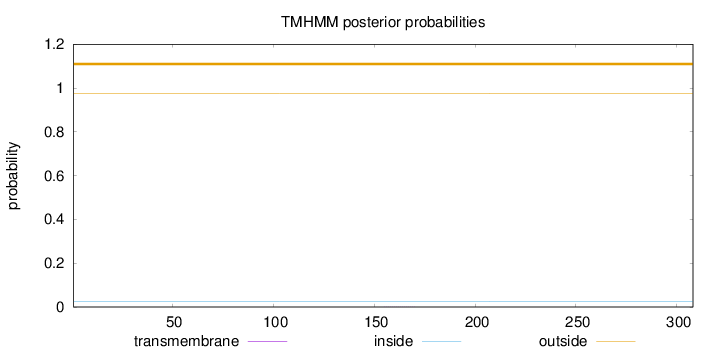
Topology
Subcellular location
Mitochondrion
Length:
308
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00233
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02503
outside
1 - 308
Population Genetic Test Statistics
Pi
315.421231
Theta
241.269133
Tajima's D
0.995917
CLR
0
CSRT
0.654417279136043
Interpretation
Uncertain
