Pre Gene Modal
BGIBMGA003119
Annotation
PREDICTED:_villin-like_protein_quail_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.631
Sequence
CDS
ATGACAACCAACTTAGAAATTAAGGGCAAGGAGTTAAGAGAAGAGGCGATTAAGCACGAAGCCAGCACAGAGCTGTTACCTCGCACACACTATGGGACATTCTATGACGCTGAAGCATACTTGGTTTACTCGGCGTCTCTTCTCTGCCATCCGGCTGGGCCGGACGTCATACGTCGAGAGATTAAAGAGAGTAATGGAGATTATGCGGAACGCCACATACACGCCTGGGCTGGAGAACGCTGCGGAAATCTTGCCGCACTCGCCCTTCGCCGTGGAACACAGCTCGCTACGTATCTTGGCATGCCTGTAGTCATTCATCGGGAGGCGTCTACCAAGGAAACTCCTCGGCTCCTTTCCTACTTTCGAGACGGTATAAGGATCCTTCGTAGCGGATCGAACTTGAATGGTGCAGCACGTTTATACCGTGTTCGTGGTCGTCGACCGCTTCTCGTCCAATTGGAACCGGTATCATGGTCTCAGTTGGCTGCAGACGGTGTATTCGTTTTAGACACCTCTTCCCTGCTCGTTCTGTGGCTGGGGCGAGCTGCCAATCTTGTCGAGAAAATATTCGGTGCCAAGATCGCTTACCGTATGGCCCGAGGAGCTGATAAAAGTACGTCAGTTCGGCGTATTGCCATCGCTCATGACGGGTACGAGCAAACTTTACCATCAAACGATCGCGCCCTACTTGACACCCTGCTAGAATTGCGAGCACGCTCAGTCCGTCCAGCTTCCCCTCCCCCCGATCCTCCTCGTACCGCTCGTCTGTACAAGGCGACACAACCTCACAGAGTTACACCTGTCACTGTACCTTCCCTAAGACCAGCAGCGAGGCTCGAAGAAGTCAAACGAGCACCGTTGTTCAGAGAAGACCTCACTGATGATGGTGTATATATAGTGGATAGTGGAAGCCGAGGAGTCTGGGCATGGATAGGACCACAAGCGACAGGTCCTGGTAAGCGTGGTGCTTTGGCAGCTGCCCGCGGCCTTGCTCGTGCTAAGCGCCACACCGGCGCAGTATGTGTGACCACAGCTGGACGAGAACCATTAGAATTTGCTTCTCTATTCCGAAATTGGCGCTGGTGTGACGCTCGACGGGATATCAGAGTTCGTGCAGCCCGCTCGGCAACTACAAAGCTGGACGCGGTTAACCTCGCCACTAACGCTTGGCTTGCTGCTGAGGCACAACTACCTGACGATGGATCGGGTTCTCTACGTGTGTGGCGCGTCCGAAGATGCGGTGTAGATGAAGACGCGGAGAAAGAGGCAGACGGTGCACTGGCAGAGGTCGAGCGACCACAAGCAGCTGCCTTTTACGATAGAGACTGTTACATTGTACTGTACACATACCATTCTCCTAACGGAGACCAAACGATGATTTACTACTGGATGGTGAGTAATTTCCTTTTTTTTATTGTTTAG
Protein
MTTNLEIKGKELREEAIKHEASTELLPRTHYGTFYDAEAYLVYSASLLCHPAGPDVIRREIKESNGDYAERHIHAWAGERCGNLAALALRRGTQLATYLGMPVVIHREASTKETPRLLSYFRDGIRILRSGSNLNGAARLYRVRGRRPLLVQLEPVSWSQLAADGVFVLDTSSLLVLWLGRAANLVEKIFGAKIAYRMARGADKSTSVRRIAIAHDGYEQTLPSNDRALLDTLLELRARSVRPASPPPDPPRTARLYKATQPHRVTPVTVPSLRPAARLEEVKRAPLFREDLTDDGVYIVDSGSRGVWAWIGPQATGPGKRGALAAARGLARAKRHTGAVCVTTAGREPLEFASLFRNWRWCDARRDIRVRAARSATTKLDAVNLATNAWLAAEAQLPDDGSGSLRVWRVRRCGVDEDAEKEADGALAEVERPQAAAFYDRDCYIVLYTYHSPNGDQTMIYYWMVSNFLFFIV
Summary
Uniprot
A0A2A4JQK9
A0A2H1VDJ5
S4P9U5
A0A212FHQ0
A0A194PNQ6
A0A194R3Y3
+ More
A0A0L7LFK3 D6WNZ0 A0A2J7PVB4 A0A2J7PVA3 A0A2J7PVB8 A0A0P5PGI0 E9FXU5 A0A0P5CMT6 A0A0P5UM74 A0A0N8BUG8 A0A0P5PY04 A0A0P5M5H3 A0A0P5NKE2 A0A0P5KT58 A0A0P5ABA6 A0A0P5SY09 A0A0P5TBZ2 A0A0P5U680 A0A0P5IPN1 A0A0N8D3X0 A0A0P5LEZ2 A0A0P5Q2N4 A0A0P5KZL2 A0A0P5KV25 A0A0P4WRH1 A0A0P5WTD3 A0A0P6H9M9 A0A0P5TXU6 A0A162REA6 A0A0P5BMB0 A0A0P5BG61 A0A0P5BF49 A0A0N8D4H7 A0A0P5XNS5 A0A0P6BQH7 A0A0P5MMQ2 A0A0P5T629 A0A0P5G4S3 A0A067RT41 A0A0P5E7T5 U4TTG0 A0A0P4WWH6 A0A1D2NEY6 A0A0P5H0X5 N6UDQ7 A0A1B6EVX4 A0A1B6F7X6 A0A0P6G4W2 A0A0P5KDN3 A0A0P5IXV7 A0A1W7R9A5 A0A0P5PSN5 A0A0P5Q5C5 A0A1B6E6N4 A0A0P5RP46 A0A1D1VB06 A0A323TSE4 A0A1B6IEW7 A0A1B6JII3 A0A1B6L7F4 A0A1B6MT39 A0A0P5H9J0 A0A0P5ZZ39 T1K4C1 A0A1Y1N463 A0A226F7B0 V5HKG4 T1IV90 A0A069DZK6 A0A0V0G6F5 A0A182G601 A0A2T7PVG9 A0A1S4EVJ4 Q17PI4 A0A0K8SQV3 A0A1W0WVC0 A0A2P8ZJT1 T1HUL6 A0A1Q3FRL2 B0WC23 A0A0L0C4D0 A0A182WHB1 A0A182MSC4 A0A182TCV7 Q7PVN2 A0A182KU95 A0A1S4H194 A0A182VMF1 A0A182Y9X0 A0A0R3NZQ9 A0A084VY72 Q29LB5
A0A0L7LFK3 D6WNZ0 A0A2J7PVB4 A0A2J7PVA3 A0A2J7PVB8 A0A0P5PGI0 E9FXU5 A0A0P5CMT6 A0A0P5UM74 A0A0N8BUG8 A0A0P5PY04 A0A0P5M5H3 A0A0P5NKE2 A0A0P5KT58 A0A0P5ABA6 A0A0P5SY09 A0A0P5TBZ2 A0A0P5U680 A0A0P5IPN1 A0A0N8D3X0 A0A0P5LEZ2 A0A0P5Q2N4 A0A0P5KZL2 A0A0P5KV25 A0A0P4WRH1 A0A0P5WTD3 A0A0P6H9M9 A0A0P5TXU6 A0A162REA6 A0A0P5BMB0 A0A0P5BG61 A0A0P5BF49 A0A0N8D4H7 A0A0P5XNS5 A0A0P6BQH7 A0A0P5MMQ2 A0A0P5T629 A0A0P5G4S3 A0A067RT41 A0A0P5E7T5 U4TTG0 A0A0P4WWH6 A0A1D2NEY6 A0A0P5H0X5 N6UDQ7 A0A1B6EVX4 A0A1B6F7X6 A0A0P6G4W2 A0A0P5KDN3 A0A0P5IXV7 A0A1W7R9A5 A0A0P5PSN5 A0A0P5Q5C5 A0A1B6E6N4 A0A0P5RP46 A0A1D1VB06 A0A323TSE4 A0A1B6IEW7 A0A1B6JII3 A0A1B6L7F4 A0A1B6MT39 A0A0P5H9J0 A0A0P5ZZ39 T1K4C1 A0A1Y1N463 A0A226F7B0 V5HKG4 T1IV90 A0A069DZK6 A0A0V0G6F5 A0A182G601 A0A2T7PVG9 A0A1S4EVJ4 Q17PI4 A0A0K8SQV3 A0A1W0WVC0 A0A2P8ZJT1 T1HUL6 A0A1Q3FRL2 B0WC23 A0A0L0C4D0 A0A182WHB1 A0A182MSC4 A0A182TCV7 Q7PVN2 A0A182KU95 A0A1S4H194 A0A182VMF1 A0A182Y9X0 A0A0R3NZQ9 A0A084VY72 Q29LB5
Pubmed
EMBL
NWSH01000819
PCG74056.1
ODYU01001971
SOQ38908.1
GAIX01003569
JAA88991.1
+ More
AGBW02008477 OWR53261.1 KQ459598 KPI94618.1 KQ460779 KPJ12234.1 JTDY01001388 KOB73996.1 KQ971343 EFA03188.1 NEVH01020965 PNF20274.1 PNF20275.1 PNF20278.1 GDIQ01128833 JAL22893.1 GL732526 EFX88163.1 GDIP01183968 JAJ39434.1 GDIP01111027 JAL92687.1 GDIQ01143715 JAL08011.1 GDIQ01128873 JAL22853.1 GDIQ01160457 JAK91268.1 GDIQ01151213 JAL00513.1 GDIQ01181094 JAK70631.1 GDIP01201517 JAJ21885.1 GDIP01136480 JAL67234.1 GDIP01128572 JAL75142.1 GDIP01117571 JAL86143.1 GDIQ01211446 JAK40279.1 GDIP01071287 JAM32428.1 GDIQ01178252 JAK73473.1 GDIQ01130138 JAL21588.1 GDIQ01202741 JAK48984.1 GDIQ01179697 JAK72028.1 GDIP01251759 GDIQ01178251 JAI71642.1 JAK73474.1 GDIP01094134 JAM09581.1 GDIQ01022720 JAN72017.1 GDIQ01207438 GDIP01137929 JAK44287.1 JAL65785.1 LRGB01000190 KZS20442.1 GDIP01182564 JAJ40838.1 GDIP01185214 JAJ38188.1 GDIP01185213 JAJ38189.1 GDIP01069631 JAM34084.1 GDIP01069630 JAM34085.1 GDIP01011366 JAM92349.1 GDIQ01154159 JAK97566.1 GDIP01132786 JAL70928.1 GDIQ01247286 JAK04439.1 KK852432 KDR23980.1 GDIP01145528 JAJ77874.1 KB630901 KB631062 ERL83940.1 ERL84052.1 GDIP01254056 JAI69345.1 LJIJ01000063 ODN03813.1 GDIP01212070 GDIQ01234833 JAJ11332.1 JAK16892.1 APGK01032343 APGK01032344 KB740839 ENN78761.1 GECZ01027916 JAS41853.1 GECZ01023469 JAS46300.1 GDIQ01040901 JAN53836.1 GDIQ01211445 JAK40280.1 GDIQ01207437 GDIP01087132 JAK44288.1 JAM16583.1 GFAH01000669 JAV47720.1 GDIQ01124116 JAL27610.1 GDIQ01125920 JAL25806.1 GEDC01003698 JAS33600.1 GDIQ01098031 JAL53695.1 BDGG01000005 GAU98834.1 MDVM02000013 PYZ99377.1 GECU01022217 JAS85489.1 GECU01008702 JAS99004.1 GEBQ01020553 JAT19424.1 GEBQ01000875 JAT39102.1 GDIQ01229532 JAK22193.1 GDIP01049929 JAM53786.1 CAEY01001566 GEZM01013353 JAV92629.1 LNIX01000001 OXA65210.1 GANP01010525 JAB73943.1 JH431578 GBGD01000475 JAC88414.1 GECL01002852 JAP03272.1 JXUM01147004 KQ570279 KXJ68399.1 PZQS01000001 PVD37426.1 CH477191 EAT48633.1 GBRD01010062 JAG55762.1 MTYJ01000042 OQV19146.1 PYGN01000035 PSN56753.1 ACPB03010465 GFDL01004838 JAV30207.1 DS231884 EDS43083.1 JRES01000934 KNC27086.1 AXCM01000590 AAAB01008984 EAA14915.4 CH379060 KRT04512.1 ATLV01018323 KE525231 KFB42916.1 EAL34130.2
AGBW02008477 OWR53261.1 KQ459598 KPI94618.1 KQ460779 KPJ12234.1 JTDY01001388 KOB73996.1 KQ971343 EFA03188.1 NEVH01020965 PNF20274.1 PNF20275.1 PNF20278.1 GDIQ01128833 JAL22893.1 GL732526 EFX88163.1 GDIP01183968 JAJ39434.1 GDIP01111027 JAL92687.1 GDIQ01143715 JAL08011.1 GDIQ01128873 JAL22853.1 GDIQ01160457 JAK91268.1 GDIQ01151213 JAL00513.1 GDIQ01181094 JAK70631.1 GDIP01201517 JAJ21885.1 GDIP01136480 JAL67234.1 GDIP01128572 JAL75142.1 GDIP01117571 JAL86143.1 GDIQ01211446 JAK40279.1 GDIP01071287 JAM32428.1 GDIQ01178252 JAK73473.1 GDIQ01130138 JAL21588.1 GDIQ01202741 JAK48984.1 GDIQ01179697 JAK72028.1 GDIP01251759 GDIQ01178251 JAI71642.1 JAK73474.1 GDIP01094134 JAM09581.1 GDIQ01022720 JAN72017.1 GDIQ01207438 GDIP01137929 JAK44287.1 JAL65785.1 LRGB01000190 KZS20442.1 GDIP01182564 JAJ40838.1 GDIP01185214 JAJ38188.1 GDIP01185213 JAJ38189.1 GDIP01069631 JAM34084.1 GDIP01069630 JAM34085.1 GDIP01011366 JAM92349.1 GDIQ01154159 JAK97566.1 GDIP01132786 JAL70928.1 GDIQ01247286 JAK04439.1 KK852432 KDR23980.1 GDIP01145528 JAJ77874.1 KB630901 KB631062 ERL83940.1 ERL84052.1 GDIP01254056 JAI69345.1 LJIJ01000063 ODN03813.1 GDIP01212070 GDIQ01234833 JAJ11332.1 JAK16892.1 APGK01032343 APGK01032344 KB740839 ENN78761.1 GECZ01027916 JAS41853.1 GECZ01023469 JAS46300.1 GDIQ01040901 JAN53836.1 GDIQ01211445 JAK40280.1 GDIQ01207437 GDIP01087132 JAK44288.1 JAM16583.1 GFAH01000669 JAV47720.1 GDIQ01124116 JAL27610.1 GDIQ01125920 JAL25806.1 GEDC01003698 JAS33600.1 GDIQ01098031 JAL53695.1 BDGG01000005 GAU98834.1 MDVM02000013 PYZ99377.1 GECU01022217 JAS85489.1 GECU01008702 JAS99004.1 GEBQ01020553 JAT19424.1 GEBQ01000875 JAT39102.1 GDIQ01229532 JAK22193.1 GDIP01049929 JAM53786.1 CAEY01001566 GEZM01013353 JAV92629.1 LNIX01000001 OXA65210.1 GANP01010525 JAB73943.1 JH431578 GBGD01000475 JAC88414.1 GECL01002852 JAP03272.1 JXUM01147004 KQ570279 KXJ68399.1 PZQS01000001 PVD37426.1 CH477191 EAT48633.1 GBRD01010062 JAG55762.1 MTYJ01000042 OQV19146.1 PYGN01000035 PSN56753.1 ACPB03010465 GFDL01004838 JAV30207.1 DS231884 EDS43083.1 JRES01000934 KNC27086.1 AXCM01000590 AAAB01008984 EAA14915.4 CH379060 KRT04512.1 ATLV01018323 KE525231 KFB42916.1 EAL34130.2
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
UP000007266
+ More
UP000235965 UP000000305 UP000076858 UP000027135 UP000030742 UP000094527 UP000019118 UP000186922 UP000247427 UP000015104 UP000198287 UP000069940 UP000249989 UP000245119 UP000008820 UP000245037 UP000015103 UP000002320 UP000037069 UP000075920 UP000075883 UP000075902 UP000007062 UP000075882 UP000075903 UP000076408 UP000001819 UP000030765
UP000235965 UP000000305 UP000076858 UP000027135 UP000030742 UP000094527 UP000019118 UP000186922 UP000247427 UP000015104 UP000198287 UP000069940 UP000249989 UP000245119 UP000008820 UP000245037 UP000015103 UP000002320 UP000037069 UP000075920 UP000075883 UP000075902 UP000007062 UP000075882 UP000075903 UP000076408 UP000001819 UP000030765
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JQK9
A0A2H1VDJ5
S4P9U5
A0A212FHQ0
A0A194PNQ6
A0A194R3Y3
+ More
A0A0L7LFK3 D6WNZ0 A0A2J7PVB4 A0A2J7PVA3 A0A2J7PVB8 A0A0P5PGI0 E9FXU5 A0A0P5CMT6 A0A0P5UM74 A0A0N8BUG8 A0A0P5PY04 A0A0P5M5H3 A0A0P5NKE2 A0A0P5KT58 A0A0P5ABA6 A0A0P5SY09 A0A0P5TBZ2 A0A0P5U680 A0A0P5IPN1 A0A0N8D3X0 A0A0P5LEZ2 A0A0P5Q2N4 A0A0P5KZL2 A0A0P5KV25 A0A0P4WRH1 A0A0P5WTD3 A0A0P6H9M9 A0A0P5TXU6 A0A162REA6 A0A0P5BMB0 A0A0P5BG61 A0A0P5BF49 A0A0N8D4H7 A0A0P5XNS5 A0A0P6BQH7 A0A0P5MMQ2 A0A0P5T629 A0A0P5G4S3 A0A067RT41 A0A0P5E7T5 U4TTG0 A0A0P4WWH6 A0A1D2NEY6 A0A0P5H0X5 N6UDQ7 A0A1B6EVX4 A0A1B6F7X6 A0A0P6G4W2 A0A0P5KDN3 A0A0P5IXV7 A0A1W7R9A5 A0A0P5PSN5 A0A0P5Q5C5 A0A1B6E6N4 A0A0P5RP46 A0A1D1VB06 A0A323TSE4 A0A1B6IEW7 A0A1B6JII3 A0A1B6L7F4 A0A1B6MT39 A0A0P5H9J0 A0A0P5ZZ39 T1K4C1 A0A1Y1N463 A0A226F7B0 V5HKG4 T1IV90 A0A069DZK6 A0A0V0G6F5 A0A182G601 A0A2T7PVG9 A0A1S4EVJ4 Q17PI4 A0A0K8SQV3 A0A1W0WVC0 A0A2P8ZJT1 T1HUL6 A0A1Q3FRL2 B0WC23 A0A0L0C4D0 A0A182WHB1 A0A182MSC4 A0A182TCV7 Q7PVN2 A0A182KU95 A0A1S4H194 A0A182VMF1 A0A182Y9X0 A0A0R3NZQ9 A0A084VY72 Q29LB5
A0A0L7LFK3 D6WNZ0 A0A2J7PVB4 A0A2J7PVA3 A0A2J7PVB8 A0A0P5PGI0 E9FXU5 A0A0P5CMT6 A0A0P5UM74 A0A0N8BUG8 A0A0P5PY04 A0A0P5M5H3 A0A0P5NKE2 A0A0P5KT58 A0A0P5ABA6 A0A0P5SY09 A0A0P5TBZ2 A0A0P5U680 A0A0P5IPN1 A0A0N8D3X0 A0A0P5LEZ2 A0A0P5Q2N4 A0A0P5KZL2 A0A0P5KV25 A0A0P4WRH1 A0A0P5WTD3 A0A0P6H9M9 A0A0P5TXU6 A0A162REA6 A0A0P5BMB0 A0A0P5BG61 A0A0P5BF49 A0A0N8D4H7 A0A0P5XNS5 A0A0P6BQH7 A0A0P5MMQ2 A0A0P5T629 A0A0P5G4S3 A0A067RT41 A0A0P5E7T5 U4TTG0 A0A0P4WWH6 A0A1D2NEY6 A0A0P5H0X5 N6UDQ7 A0A1B6EVX4 A0A1B6F7X6 A0A0P6G4W2 A0A0P5KDN3 A0A0P5IXV7 A0A1W7R9A5 A0A0P5PSN5 A0A0P5Q5C5 A0A1B6E6N4 A0A0P5RP46 A0A1D1VB06 A0A323TSE4 A0A1B6IEW7 A0A1B6JII3 A0A1B6L7F4 A0A1B6MT39 A0A0P5H9J0 A0A0P5ZZ39 T1K4C1 A0A1Y1N463 A0A226F7B0 V5HKG4 T1IV90 A0A069DZK6 A0A0V0G6F5 A0A182G601 A0A2T7PVG9 A0A1S4EVJ4 Q17PI4 A0A0K8SQV3 A0A1W0WVC0 A0A2P8ZJT1 T1HUL6 A0A1Q3FRL2 B0WC23 A0A0L0C4D0 A0A182WHB1 A0A182MSC4 A0A182TCV7 Q7PVN2 A0A182KU95 A0A1S4H194 A0A182VMF1 A0A182Y9X0 A0A0R3NZQ9 A0A084VY72 Q29LB5
PDB
3FFN
E-value=1.55798e-20,
Score=246
Ontologies
KEGG
GO
PANTHER
Topology
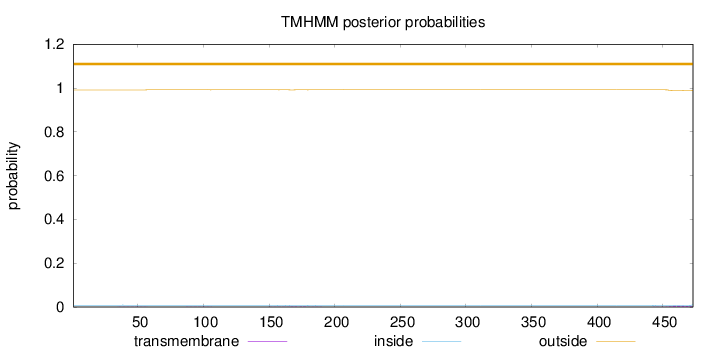
Length:
473
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13309
Exp number, first 60 AAs:
0.00773
Total prob of N-in:
0.00856
outside
1 - 473
Population Genetic Test Statistics
Pi
174.437546
Theta
193.891151
Tajima's D
-0.831675
CLR
10.22345
CSRT
0.170441477926104
Interpretation
Uncertain
