Pre Gene Modal
BGIBMGA003038
Annotation
PREDICTED:_rho-associated_protein_kinase_isoform_X1_[Bombyx_mori]
Full name
Rho-associated protein kinase
Location in the cell
Nuclear Reliability : 2.971
Sequence
CDS
ATGAAGGATTGCCTTAGTGGCCACAGAGATCAAGAACACAACAGGATGGAAGTGAAGGATGAGGATCGCATGAGGCGTCTTCATGCGTTGGAGGAACGTTTGTGTGACCCCCGTTCCACCGGTAATGTGGACTGTCTACTGGATACCGTCACTGCGCTCGTGTCCGATTGTGACCATCCAGCTATTCGTCGCATGAAAAACGTCGAAGCCTACACTAGTAGATATGAAGAGTTTTCTTCAGAAATTATTAATCTGCGAATGAAGGCAGCTGATTTCCATTTAATTAAAGTGATTGGACGAGGGGCATTTGGTGAAGTACAACTAGTCAGACAAAAGTCTACCAACCATGTTTATGCCATGAAACTTCTTAGTAAAGTAGAAATGATTAAGAGGTCAGACTCCACATTCTTTTGGGAAGAAAGGCACATTATGGCCCATGCCAACTCGGACTGGATACTAAAATTACATTTTGCATTTCAAGATCATAAATATCTTTATATGGTTATGGATTATATGCCTGGTGGTGATCTTGTCAGTCTAATGTCGAACTATGACATACCAGAGAAATGGGCAAAATTCTACACAATGGAGATTGTTCTTGCTCTGGATGTTATTCATGGCATGGGATTTGTCCATAGAGATGTAAAACCAGACAATATGTTGATAGATAAACATGGCCATTTAAAACTAGCCGATTTTGGTACTTGTATGAGAATGGGACTGGATGGTTTAGTTCGAGCAAGCAATGCTGTGGGAACTCCTGATTATATATCTCCTGAAGTTCTGCAGTCACAAAATGGTGAAGGAGTATATGGGCGTGAGTGCGATTGGTGGGCAGTAGGAATATTTTTGTACGAAATGCTTATTGGTGACCCTCCTTTTTATGCTGATAGTTTAGTGGGCACGTATGGTAAAATTATGGATCACAGGAATTCTTTACAGTTTCCTGATGATGTTGAGATCTCAAAAGAAGCAAAGTCTCTAATTAGAGGACTTTTAACTGATAGAACTAAAAGATTAGGTAGAAACAGTGTGGATGAAATTAAACAGCATCCATTTTTTATAAATGATCAATGGAGTTTTGAAAATCTGAGGGATTCTGTACCTCCTGTGGTGCCTGAACTCTCCAGTGATGATGATACTAGGAACTTTGATGATATTGAAAAATCAGATGCACTTGATGAATCCTTTCCTGTACCAAAAGCATTTGTTGGTAATCATTTGCCATTTGTTGGTTTCACTTATAATGGTGATTACCAATTGTGCACTCGCCAAAAGAAAGCAAATGACGTAGTAGACACAATATCTAATAATCATATCAACAATGATGGATCTGAAGCCATATATCAACTAGAGAAACTGCTCGAAAGAGAAAGAGATGGCAAACGTAAACTAGAAGATACTCAAGCTGCCCTATGTGCTCAGCTCGAAGAACTATCACAGAGAGAACTACGCAATAAAAAAATAATTTCTGAAAGCGATAAAGAGGTTGCATTACTGAGACATGATCTTAAAGAAATTCAACGGATAGCTGAACTGGAAGTAGAATCTAGACGAAAGGCTGAAGCTAATCTCAATGAGGCAAAACGCCGTCTTGAAGAGGAACAAACCAAGAGAACGAAAGAAATGAGTAATTTACACATATACAACGAAAAAATTAACGCATTAGAAAAACAGCTCGAAGAATTGCGAGAGAAACTAAAACAAGAGTCTGAAGCAGCAGCCAAATCTAGGAAGCAAGCTGCCGAGCTATCGGCTGCCCAGGCCGCCGCCGCAGCTGTCAACGACGGCACGGTAACCAGCTTGCGGGCTCAACGTGATGCTCTCGAACGAGAGAGAAGTATTTTATCAGAGGAACTGAGCGAAGTTAAGGCAGCTAGACAACGTTCGGAAGCGGCGGTCACAGAGGCCACCTGCAGACTCAATGCGGCTCATGCAGAATTGGAGCGCACTTCGACACGATTATCTCAAATGATCGCTGATAACAGACAATTAACAGAACGTGTATCTTCACTAGAGAAGGAATGCACTTCATTATCTCATGAGTTAAGATCAGCGCAACATTTATATCAACAAGAGTTGCGTGCACATCAGGACACGCAACGATCTCAGCTTCTCTCTAAACAAGAAGCCAACTTGGAACTAGTCAAGACCCTGCAAACTAAGCTGAACGAAGAAAAGACTGCCAGACAACGGTCGGAATCAGCTTGTCAAGAGAAAGACCGACAGATGTCGATGCTCAGCGTTGATTACAGACAGATGCAGCAGAGACTCCAGAAACTAGAGGGCGAACATCGGCAAGAAAGCGAAAAGGTATGCGGGCTGGTCGCGTCGCTAGAGCAGGAGCGCGCGGCTCGACTGGCGGCGTCGGGCGAGGTGTCGGCGGCGGAGGCGGCGGCCCGCACCGCGCTCGCCGAGCGGGACGCGCGCGCCCGCGACCTGCACGCCGTGCGCGCCGCGCTGCACGACGCCTCCGAGAAACTAGCCGCCGCCAGCGCAGAGCGGGACATGTACTACGCCAGGAGTGAAGAGTTGCGCTCTCAGTTGGAGAATGAGCAACATTACTGCCATGTGTATAGAAGTCAGGTCAATGAGATCCGCACTCAGGTGGAGGAGAGTTCTCGCGCTGCCCGGGACCTCGAACAAGAAAGATCCAGCTTGTTGCATCAACTACAGCTCGCCATTGCTAGGGCCGACTCTGAGGCTATTGCAAGATCGATAGCGGAAGAGACAGTTGGTGAACTGGAGAAGGAGAAGACAATGAAAGAGTTAGAGATGCGGGAAGCTGTGTCCCAGCATCGCGCCGAGCTGGCGGCCCGCGACAACATGCTGCAGGGCCTGCGGGACCGGGACCACGAGAACCGCGCCACCATCGACCTGCAGCGGAAGGAGGTGGACGAGCTCCGGCGCAGCGGCACGGCGCTGGCGGAGCGCGTGGCGTCGCTGCAGCGGCTGCAGGACGAGCTGGACCGACTCAACAAGAAGCTCAACTCTGAGATCATGCTCAAGCAGCAGGCGGTCAACAAGCTCGCCGAGATCATGAACAGGAAAGACATGAACCCGACTGCGGGCAAGAACAAGAGCAAGATGTCGGTCCGGAAGGACAAGGATTACCGCAAGCTGCAGCAAGAGCTGACCCGGGAGAAGGAGAAGTTCGACCAGCACATCTCGAAGCTGCAGCGGGACCTGCAGGACTGTCAGCAGCAGTTGCTGGACGAGCAGTCCACCAGGCTTAGACTTGCCATGGAGGTTGACAGCAAAGACGCTGAGATCGAGCAGCTGAAGGAGAAGCTCGCCGCGCTCACCAGCGAGACGGCGTCGCAGTCGTCGGCCGACGCGGAGGACGGCGAGAACGAGCAGATCCTCGAAGGCTGGCTCTCGGTGCCGTTCAAGCAGAACATACGCAGGCACGGCTGGAAGAAGCAGTACGTCGTCGTTTCCTCCAAGAAGATCATCTTCTACAACTCTGAGAACGATAAACAGAATACCACAGATCCCGTCATGATCTTGGATTTGAGCAAGGTGTTCCACGTGCGCTCCGTGACGCAGGGCGACGTGATCCGCGCCGACGCGAAGGACATCCCGCGGATCTTCCAGCTGCTGTACGCGGGCGAGGGGGAGGCGCGCCGCCCCGACCAGCAGGACCAGCCGCAGGACTCCACCGACCATCAAGGCAACACGGTGCAGCACAAAGGCCACGACCTGGTGAGCATCACGTACCACATCCCGACGGCGTGCGAGGTGTGCACGCGGCCGCTGTGGCACATGTTCCGGCCGCCGCAGGCCTACGAGTGTCGCCGGTGTCGGATGAAGATCCACGCGGAGCACCTGACGGAGGGCGAGGGCGTGGCGGCGTGCAAGCTGCACGCGGACCGCGCGCGCGAGCTGCTGCTGCTGGCGCCCGCCGCGCCCGAGCAGCGCCGCTGGGTGGCCCGCCTGGCGCGCCGCGTGCAGCGACACGGCTACCGCGCCGCGCACCACAACCACGACCATCGCCTCTCGCCCAGGGACTCTATGCGCTCGAACCTGAAGTCGGCCGCGCACCTGAACGTGTCCCAGCGCAGCTCCACGCTGCCCGCCAACGCGTCCATGCAGCGGCAGTGA
Protein
MKDCLSGHRDQEHNRMEVKDEDRMRRLHALEERLCDPRSTGNVDCLLDTVTALVSDCDHPAIRRMKNVEAYTSRYEEFSSEIINLRMKAADFHLIKVIGRGAFGEVQLVRQKSTNHVYAMKLLSKVEMIKRSDSTFFWEERHIMAHANSDWILKLHFAFQDHKYLYMVMDYMPGGDLVSLMSNYDIPEKWAKFYTMEIVLALDVIHGMGFVHRDVKPDNMLIDKHGHLKLADFGTCMRMGLDGLVRASNAVGTPDYISPEVLQSQNGEGVYGRECDWWAVGIFLYEMLIGDPPFYADSLVGTYGKIMDHRNSLQFPDDVEISKEAKSLIRGLLTDRTKRLGRNSVDEIKQHPFFINDQWSFENLRDSVPPVVPELSSDDDTRNFDDIEKSDALDESFPVPKAFVGNHLPFVGFTYNGDYQLCTRQKKANDVVDTISNNHINNDGSEAIYQLEKLLERERDGKRKLEDTQAALCAQLEELSQRELRNKKIISESDKEVALLRHDLKEIQRIAELEVESRRKAEANLNEAKRRLEEEQTKRTKEMSNLHIYNEKINALEKQLEELREKLKQESEAAAKSRKQAAELSAAQAAAAAVNDGTVTSLRAQRDALERERSILSEELSEVKAARQRSEAAVTEATCRLNAAHAELERTSTRLSQMIADNRQLTERVSSLEKECTSLSHELRSAQHLYQQELRAHQDTQRSQLLSKQEANLELVKTLQTKLNEEKTARQRSESACQEKDRQMSMLSVDYRQMQQRLQKLEGEHRQESEKVCGLVASLEQERAARLAASGEVSAAEAAARTALAERDARARDLHAVRAALHDASEKLAAASAERDMYYARSEELRSQLENEQHYCHVYRSQVNEIRTQVEESSRAARDLEQERSSLLHQLQLAIARADSEAIARSIAEETVGELEKEKTMKELEMREAVSQHRAELAARDNMLQGLRDRDHENRATIDLQRKEVDELRRSGTALAERVASLQRLQDELDRLNKKLNSEIMLKQQAVNKLAEIMNRKDMNPTAGKNKSKMSVRKDKDYRKLQQELTREKEKFDQHISKLQRDLQDCQQQLLDEQSTRLRLAMEVDSKDAEIEQLKEKLAALTSETASQSSADAEDGENEQILEGWLSVPFKQNIRRHGWKKQYVVVSSKKIIFYNSENDKQNTTDPVMILDLSKVFHVRSVTQGDVIRADAKDIPRIFQLLYAGEGEARRPDQQDQPQDSTDHQGNTVQHKGHDLVSITYHIPTACEVCTRPLWHMFRPPQAYECRRCRMKIHAEHLTEGEGVAACKLHADRARELLLLAPAAPEQRRWVARLARRVQRHGYRAAHHNHDHRLSPRDSMRSNLKSAAHLNVSQRSSTLPANASMQRQ
Summary
Description
Protein kinase which is a key regulator of actin cytoskeleton and cell polarity.
Catalytic Activity
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Subunit
Homodimer.
Similarity
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family.
Uniprot
E3VQ36
A0A2A4JQS9
A0A2H1VDL0
A0A194R8N7
A0A194PPF9
A0A0L7LEM1
+ More
A0A2J7PRJ7 A0A2J7PRI0 A0A151XBB6 E2B7A8 A0A154NXD5 F4WIC6 A0A088ATG8 A0A2J7PRI1 A0A195BLB5 A0A158NZV1 A0A0L7RA01 A0A3L8DV67 A0A195F385 A0A151J2D9 D6WNY9 A0A026WPN6 A0A1Y1M4Z7 E2A893 A0A232EM27 K7IV18 A0A310SKS9 A0A2A3EID4 A0A0C9QM01 A0A195CM24 A0A139WHG7 A0A0C9QED7 A0A1Y1LZZ6 E0VY20 A0A023F4N1 A0A0C9R4B6 A0A2S2NMH7 A0A2S2Q665 A0A0C9R4C1 J9JZB8 A0A067RLR3 A0A224X567 A0A146KUB8 T1HUL2 A0A1S4G7D4 A0A0K8WJ47 A0A0L0CL34 B0WWT9 W8BVK1 A0A0A1WEU6 A0A1Q3F3X3 A0A0K8TVJ8 W8AW70 A0A1L8DM40 A0A1B0BMY5 A0A1I8Q3E9 E9GLV2 A0A182H9C4 A0A1I8Q3C7 A0A1A9XY85 A0A0P5W602 A0A1I8N3J7 A0A0N8EBV9 A0A0P5B6Y5 A0A0N8D6J6 A0A1J1HSG0 A0A0P5U236 A0A0P4ZIX4 A0A0P5WU04 A0A0P5B8Z3 A0A0P5CVD0 A0A1I8Q3B7 W4VRL9 A0A1A9VDC0 A0A0T6ATL5 A0A1I8Q3C4 A0A1I8Q3E3 A0A1I8Q3E8 A0A1I8Q3C1 A0A1A9Z8L7 A0A0P5ZFX7 A0A0P6ACA1 A0A0N8DLC4 A0A0P5CUK8 A0A1B0FAA8 A0A1A9WDB5 A0A0P6B1F2 X2JC83 A0A182FJW0 Q179S5 A0A182JLX8 A0A0Q5TJ56 Q9VXE3 B3MRC3 Q961D4 Q9U779 B3NVJ7 X2JKQ8 A0A0P8XK28 A0A1W4VJ27
A0A2J7PRJ7 A0A2J7PRI0 A0A151XBB6 E2B7A8 A0A154NXD5 F4WIC6 A0A088ATG8 A0A2J7PRI1 A0A195BLB5 A0A158NZV1 A0A0L7RA01 A0A3L8DV67 A0A195F385 A0A151J2D9 D6WNY9 A0A026WPN6 A0A1Y1M4Z7 E2A893 A0A232EM27 K7IV18 A0A310SKS9 A0A2A3EID4 A0A0C9QM01 A0A195CM24 A0A139WHG7 A0A0C9QED7 A0A1Y1LZZ6 E0VY20 A0A023F4N1 A0A0C9R4B6 A0A2S2NMH7 A0A2S2Q665 A0A0C9R4C1 J9JZB8 A0A067RLR3 A0A224X567 A0A146KUB8 T1HUL2 A0A1S4G7D4 A0A0K8WJ47 A0A0L0CL34 B0WWT9 W8BVK1 A0A0A1WEU6 A0A1Q3F3X3 A0A0K8TVJ8 W8AW70 A0A1L8DM40 A0A1B0BMY5 A0A1I8Q3E9 E9GLV2 A0A182H9C4 A0A1I8Q3C7 A0A1A9XY85 A0A0P5W602 A0A1I8N3J7 A0A0N8EBV9 A0A0P5B6Y5 A0A0N8D6J6 A0A1J1HSG0 A0A0P5U236 A0A0P4ZIX4 A0A0P5WU04 A0A0P5B8Z3 A0A0P5CVD0 A0A1I8Q3B7 W4VRL9 A0A1A9VDC0 A0A0T6ATL5 A0A1I8Q3C4 A0A1I8Q3E3 A0A1I8Q3E8 A0A1I8Q3C1 A0A1A9Z8L7 A0A0P5ZFX7 A0A0P6ACA1 A0A0N8DLC4 A0A0P5CUK8 A0A1B0FAA8 A0A1A9WDB5 A0A0P6B1F2 X2JC83 A0A182FJW0 Q179S5 A0A182JLX8 A0A0Q5TJ56 Q9VXE3 B3MRC3 Q961D4 Q9U779 B3NVJ7 X2JKQ8 A0A0P8XK28 A0A1W4VJ27
EC Number
2.7.11.1
Pubmed
19121390
21040523
26354079
26227816
20798317
21719571
+ More
21347285 30249741 18362917 19820115 24508170 28004739 28648823 20075255 20566863 25474469 24845553 26823975 17510324 26108605 24495485 25830018 21292972 26483478 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 26109357 26109356 10570971 18057021
21347285 30249741 18362917 19820115 24508170 28004739 28648823 20075255 20566863 25474469 24845553 26823975 17510324 26108605 24495485 25830018 21292972 26483478 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 26109357 26109356 10570971 18057021
EMBL
BABH01010310
BABH01010311
HQ179969
ADP21243.1
NWSH01000819
PCG74058.1
+ More
ODYU01001971 SOQ38907.1 KQ460779 KPJ12236.1 KQ459598 KPI94619.1 JTDY01001388 KOB73998.1 NEVH01021962 PNF18939.1 PNF18941.1 KQ982330 KYQ57673.1 GL446154 EFN88393.1 KQ434777 KZC04287.1 GL888175 EGI66057.1 PNF18940.1 KQ976440 KYM86493.1 ADTU01005006 ADTU01005007 KQ414621 KOC67581.1 QOIP01000003 RLU24317.1 KQ981856 KYN34642.1 KQ980404 KYN16200.1 KQ971343 EFA04406.2 KK107144 EZA57596.1 GEZM01044341 JAV78297.1 GL437497 EFN70359.1 NNAY01003440 OXU19414.1 AAZX01001198 AAZX01004095 KQ761797 OAD56794.1 KZ288253 PBC30946.1 GBYB01001662 GBYB01001665 JAG71429.1 JAG71432.1 KQ977580 KYN01758.1 KYB27402.1 GBYB01001664 JAG71431.1 GEZM01044340 JAV78298.1 DS235843 EEB18276.1 GBBI01002375 JAC16337.1 GBYB01001661 JAG71428.1 GGMR01005771 MBY18390.1 GGMS01004006 MBY73209.1 GBYB01001663 GBYB01001666 JAG71430.1 JAG71433.1 ABLF02039304 KK852432 KDR23978.1 GFTR01008781 JAW07645.1 GDHC01019414 JAP99214.1 ACPB03002620 GDHF01025419 GDHF01001138 JAI26895.1 JAI51176.1 JRES01000336 KNC32174.1 DS232152 EDS36200.1 GAMC01013376 JAB93179.1 GBXI01017086 GBXI01016730 GBXI01004869 JAC97205.1 JAC97561.1 JAD09423.1 GFDL01012826 JAV22219.1 GDHF01033817 GDHF01030591 JAI18497.1 JAI21723.1 GAMC01013375 JAB93180.1 GFDF01006541 JAV07543.1 JXJN01017103 GL732551 EFX79573.1 JXUM01029960 KQ560869 KXJ80552.1 GDIP01252860 GDIP01251110 GDIP01235514 GDIP01195962 GDIP01191210 GDIP01179469 GDIP01172122 GDIP01148219 GDIP01147865 GDIP01136323 GDIP01104174 GDIP01104173 GDIP01101173 GDIP01083627 JAL99541.1 GDIQ01041996 JAN52741.1 GDIP01188611 JAJ34791.1 GDIP01212269 GDIP01181364 GDIP01166213 GDIP01122721 GDIP01120878 GDIP01087678 GDIP01086633 GDIP01085230 GDIP01079325 GDIP01072826 GDIP01063879 GDIP01058984 GDIP01058983 GDIP01039587 GDIP01038536 GDIP01037388 JAM39836.1 CVRI01000020 CRK90991.1 GDIP01118832 JAL84882.1 GDIP01216010 JAJ07392.1 GDIP01081777 JAM21938.1 GDIP01203260 JAJ20142.1 GDIP01165018 JAJ58384.1 GANO01001708 JAB58163.1 LJIG01022848 KRT78409.1 GDIP01045934 JAM57781.1 GDIP01036387 JAM67328.1 GDIP01022455 JAM81260.1 GDIP01166214 JAJ57188.1 CCAG010017193 GDIP01035245 JAM68470.1 AE014298 AHN59805.1 CH477346 EAT42995.1 CH954180 KQS29958.1 BT044181 AAF48631.1 ACH92246.1 CH902622 EDV34328.1 AY051659 AAK93083.1 AF151375 AAF03776.1 EDV46389.2 AHN59807.1 KPU75083.1 KPU75084.1 KPU75085.1
ODYU01001971 SOQ38907.1 KQ460779 KPJ12236.1 KQ459598 KPI94619.1 JTDY01001388 KOB73998.1 NEVH01021962 PNF18939.1 PNF18941.1 KQ982330 KYQ57673.1 GL446154 EFN88393.1 KQ434777 KZC04287.1 GL888175 EGI66057.1 PNF18940.1 KQ976440 KYM86493.1 ADTU01005006 ADTU01005007 KQ414621 KOC67581.1 QOIP01000003 RLU24317.1 KQ981856 KYN34642.1 KQ980404 KYN16200.1 KQ971343 EFA04406.2 KK107144 EZA57596.1 GEZM01044341 JAV78297.1 GL437497 EFN70359.1 NNAY01003440 OXU19414.1 AAZX01001198 AAZX01004095 KQ761797 OAD56794.1 KZ288253 PBC30946.1 GBYB01001662 GBYB01001665 JAG71429.1 JAG71432.1 KQ977580 KYN01758.1 KYB27402.1 GBYB01001664 JAG71431.1 GEZM01044340 JAV78298.1 DS235843 EEB18276.1 GBBI01002375 JAC16337.1 GBYB01001661 JAG71428.1 GGMR01005771 MBY18390.1 GGMS01004006 MBY73209.1 GBYB01001663 GBYB01001666 JAG71430.1 JAG71433.1 ABLF02039304 KK852432 KDR23978.1 GFTR01008781 JAW07645.1 GDHC01019414 JAP99214.1 ACPB03002620 GDHF01025419 GDHF01001138 JAI26895.1 JAI51176.1 JRES01000336 KNC32174.1 DS232152 EDS36200.1 GAMC01013376 JAB93179.1 GBXI01017086 GBXI01016730 GBXI01004869 JAC97205.1 JAC97561.1 JAD09423.1 GFDL01012826 JAV22219.1 GDHF01033817 GDHF01030591 JAI18497.1 JAI21723.1 GAMC01013375 JAB93180.1 GFDF01006541 JAV07543.1 JXJN01017103 GL732551 EFX79573.1 JXUM01029960 KQ560869 KXJ80552.1 GDIP01252860 GDIP01251110 GDIP01235514 GDIP01195962 GDIP01191210 GDIP01179469 GDIP01172122 GDIP01148219 GDIP01147865 GDIP01136323 GDIP01104174 GDIP01104173 GDIP01101173 GDIP01083627 JAL99541.1 GDIQ01041996 JAN52741.1 GDIP01188611 JAJ34791.1 GDIP01212269 GDIP01181364 GDIP01166213 GDIP01122721 GDIP01120878 GDIP01087678 GDIP01086633 GDIP01085230 GDIP01079325 GDIP01072826 GDIP01063879 GDIP01058984 GDIP01058983 GDIP01039587 GDIP01038536 GDIP01037388 JAM39836.1 CVRI01000020 CRK90991.1 GDIP01118832 JAL84882.1 GDIP01216010 JAJ07392.1 GDIP01081777 JAM21938.1 GDIP01203260 JAJ20142.1 GDIP01165018 JAJ58384.1 GANO01001708 JAB58163.1 LJIG01022848 KRT78409.1 GDIP01045934 JAM57781.1 GDIP01036387 JAM67328.1 GDIP01022455 JAM81260.1 GDIP01166214 JAJ57188.1 CCAG010017193 GDIP01035245 JAM68470.1 AE014298 AHN59805.1 CH477346 EAT42995.1 CH954180 KQS29958.1 BT044181 AAF48631.1 ACH92246.1 CH902622 EDV34328.1 AY051659 AAK93083.1 AF151375 AAF03776.1 EDV46389.2 AHN59807.1 KPU75083.1 KPU75084.1 KPU75085.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000235965
+ More
UP000075809 UP000008237 UP000076502 UP000007755 UP000005203 UP000078540 UP000005205 UP000053825 UP000279307 UP000078541 UP000078492 UP000007266 UP000053097 UP000000311 UP000215335 UP000002358 UP000242457 UP000078542 UP000009046 UP000007819 UP000027135 UP000015103 UP000037069 UP000002320 UP000092460 UP000095300 UP000000305 UP000069940 UP000249989 UP000092443 UP000095301 UP000183832 UP000078200 UP000092445 UP000092444 UP000091820 UP000000803 UP000069272 UP000008820 UP000075880 UP000008711 UP000007801 UP000192221
UP000075809 UP000008237 UP000076502 UP000007755 UP000005203 UP000078540 UP000005205 UP000053825 UP000279307 UP000078541 UP000078492 UP000007266 UP000053097 UP000000311 UP000215335 UP000002358 UP000242457 UP000078542 UP000009046 UP000007819 UP000027135 UP000015103 UP000037069 UP000002320 UP000092460 UP000095300 UP000000305 UP000069940 UP000249989 UP000092443 UP000095301 UP000183832 UP000078200 UP000092445 UP000092444 UP000091820 UP000000803 UP000069272 UP000008820 UP000075880 UP000008711 UP000007801 UP000192221
Interpro
SUPFAM
SSF56112
SSF56112
Gene 3D
CDD
ProteinModelPortal
E3VQ36
A0A2A4JQS9
A0A2H1VDL0
A0A194R8N7
A0A194PPF9
A0A0L7LEM1
+ More
A0A2J7PRJ7 A0A2J7PRI0 A0A151XBB6 E2B7A8 A0A154NXD5 F4WIC6 A0A088ATG8 A0A2J7PRI1 A0A195BLB5 A0A158NZV1 A0A0L7RA01 A0A3L8DV67 A0A195F385 A0A151J2D9 D6WNY9 A0A026WPN6 A0A1Y1M4Z7 E2A893 A0A232EM27 K7IV18 A0A310SKS9 A0A2A3EID4 A0A0C9QM01 A0A195CM24 A0A139WHG7 A0A0C9QED7 A0A1Y1LZZ6 E0VY20 A0A023F4N1 A0A0C9R4B6 A0A2S2NMH7 A0A2S2Q665 A0A0C9R4C1 J9JZB8 A0A067RLR3 A0A224X567 A0A146KUB8 T1HUL2 A0A1S4G7D4 A0A0K8WJ47 A0A0L0CL34 B0WWT9 W8BVK1 A0A0A1WEU6 A0A1Q3F3X3 A0A0K8TVJ8 W8AW70 A0A1L8DM40 A0A1B0BMY5 A0A1I8Q3E9 E9GLV2 A0A182H9C4 A0A1I8Q3C7 A0A1A9XY85 A0A0P5W602 A0A1I8N3J7 A0A0N8EBV9 A0A0P5B6Y5 A0A0N8D6J6 A0A1J1HSG0 A0A0P5U236 A0A0P4ZIX4 A0A0P5WU04 A0A0P5B8Z3 A0A0P5CVD0 A0A1I8Q3B7 W4VRL9 A0A1A9VDC0 A0A0T6ATL5 A0A1I8Q3C4 A0A1I8Q3E3 A0A1I8Q3E8 A0A1I8Q3C1 A0A1A9Z8L7 A0A0P5ZFX7 A0A0P6ACA1 A0A0N8DLC4 A0A0P5CUK8 A0A1B0FAA8 A0A1A9WDB5 A0A0P6B1F2 X2JC83 A0A182FJW0 Q179S5 A0A182JLX8 A0A0Q5TJ56 Q9VXE3 B3MRC3 Q961D4 Q9U779 B3NVJ7 X2JKQ8 A0A0P8XK28 A0A1W4VJ27
A0A2J7PRJ7 A0A2J7PRI0 A0A151XBB6 E2B7A8 A0A154NXD5 F4WIC6 A0A088ATG8 A0A2J7PRI1 A0A195BLB5 A0A158NZV1 A0A0L7RA01 A0A3L8DV67 A0A195F385 A0A151J2D9 D6WNY9 A0A026WPN6 A0A1Y1M4Z7 E2A893 A0A232EM27 K7IV18 A0A310SKS9 A0A2A3EID4 A0A0C9QM01 A0A195CM24 A0A139WHG7 A0A0C9QED7 A0A1Y1LZZ6 E0VY20 A0A023F4N1 A0A0C9R4B6 A0A2S2NMH7 A0A2S2Q665 A0A0C9R4C1 J9JZB8 A0A067RLR3 A0A224X567 A0A146KUB8 T1HUL2 A0A1S4G7D4 A0A0K8WJ47 A0A0L0CL34 B0WWT9 W8BVK1 A0A0A1WEU6 A0A1Q3F3X3 A0A0K8TVJ8 W8AW70 A0A1L8DM40 A0A1B0BMY5 A0A1I8Q3E9 E9GLV2 A0A182H9C4 A0A1I8Q3C7 A0A1A9XY85 A0A0P5W602 A0A1I8N3J7 A0A0N8EBV9 A0A0P5B6Y5 A0A0N8D6J6 A0A1J1HSG0 A0A0P5U236 A0A0P4ZIX4 A0A0P5WU04 A0A0P5B8Z3 A0A0P5CVD0 A0A1I8Q3B7 W4VRL9 A0A1A9VDC0 A0A0T6ATL5 A0A1I8Q3C4 A0A1I8Q3E3 A0A1I8Q3E8 A0A1I8Q3C1 A0A1A9Z8L7 A0A0P5ZFX7 A0A0P6ACA1 A0A0N8DLC4 A0A0P5CUK8 A0A1B0FAA8 A0A1A9WDB5 A0A0P6B1F2 X2JC83 A0A182FJW0 Q179S5 A0A182JLX8 A0A0Q5TJ56 Q9VXE3 B3MRC3 Q961D4 Q9U779 B3NVJ7 X2JKQ8 A0A0P8XK28 A0A1W4VJ27
PDB
6E9W
E-value=4.81483e-171,
Score=1548
Ontologies
GO
GO:0035556
GO:0046872
GO:0004674
GO:0017048
GO:0005524
GO:0007266
GO:0018107
GO:0031032
GO:0030866
GO:0005856
GO:0072518
GO:0005737
GO:0048598
GO:0000281
GO:0017049
GO:1901888
GO:0004697
GO:0005938
GO:0000022
GO:0008335
GO:0001736
GO:0050770
GO:2000040
GO:0006468
GO:0044291
GO:0030036
GO:0032956
GO:0007309
GO:0051020
GO:0008544
GO:0007302
GO:0016331
GO:0045184
GO:0035317
GO:0001932
GO:0032065
GO:0042325
GO:1901739
GO:0090303
GO:0031532
GO:0007391
GO:0042067
GO:0035191
GO:0003384
GO:0048599
GO:0032231
GO:0008360
GO:0007300
GO:0001737
GO:1903035
GO:0051496
GO:0000278
GO:1901648
GO:0106037
GO:0106036
GO:0005829
GO:0004672
GO:0051603
GO:0008483
GO:0006418
GO:0016507
GO:0048384
PANTHER
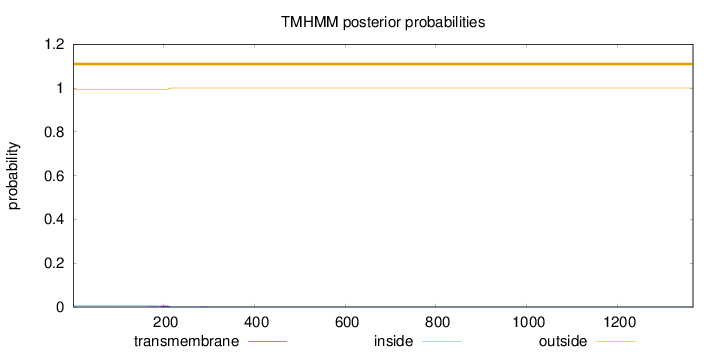
Topology
Length:
1367
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.124
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00568
outside
1 - 1367
Population Genetic Test Statistics
Pi
182.356149
Theta
173.783325
Tajima's D
0.199472
CLR
17.783966
CSRT
0.423028848557572
Interpretation
Uncertain
