Gene
KWMTBOMO02209
Pre Gene Modal
BGIBMGA003037
Annotation
PREDICTED:_cytosolic_carboxypeptidase_6_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.457
Sequence
CDS
ATGCTGAACCCCGACGGTGTATTTCTGGGCAACCAACGCGCCGATCTGTTTGGAGCCGACCTAAATCGTTCGTGGAACAGAGCTACGACCTTTGCTCATCCCGCTCTTGTTGCTGTTAACGATCTTATTAAGAAGCTTACTGCGGAGAAGGGCGTCCAACTGGACTTCATTATTGATTTACATGCTGATATAAGTCATGAAGGAGTTTTTGTTCGCGGGAATTCATATGATGACGTCTATAGGTTTGAGCGTCACGCAGTGCTACCTAAATTCTTAGGTGGTCGTATAGAAGCATGGAAAGCAGAGATTTGCCTTTACAACGCTGATCCTATCGTCGCTGGCAGCGCACGAAGAGCTCTTCCAGTTGCTGGCAACATTGACGCTTATTCAATTTTAGTATCGCTTGGTGGGAGACGGCTTAATCCCAGAGGACCTTATTTACATTACACTGAAGATGCTTGTATCCTTTTAAGAATAAGTGATTTTAGATTATGTTAG
Protein
MLNPDGVFLGNQRADLFGADLNRSWNRATTFAHPALVAVNDLIKKLTAEKGVQLDFIIDLHADISHEGVFVRGNSYDDVYRFERHAVLPKFLGGRIEAWKAEICLYNADPIVAGSARRALPVAGNIDAYSILVSLGGRRLNPRGPYLHYTEDACILLRISDFRLC
Summary
Uniprot
H9J0K0
A0A2H1VDJ8
A0A212FEH3
A0A194R3Y8
A0A194PNR1
A0A0L7LF39
+ More
A0A0L7KSF3 A0A2A4JRH4 A0A2A3EBV4 A0A087ZTQ6 A0A0L7RBK6 K7IPW7 A0A1B0DQG4 A0A0M8ZTI9 E0VBM5 A0A182UNK7 A0A182HGR8 A0A182JX26 A0A182KMA2 A7UVJ6 W5JDL5 A0A182TGL6 A0A182VQS9 A0A182XVA0 A0A182F6V7 A0A1B0CAC9 W8BAZ6 A0A182RP05 A0A182MTY9 A0A182X2P8 D6W7B0 A0A182N503 A0A182GQN1 A0A0J7NKS2 A0A336MLQ2 A0A336LD69 A0A0C9RHE3 B0W961 A0A1I8N3B3 A0A0K8W735 A0A1I8P0G1 A0A1I8P0A2 A0A1B0FRI8 A0A182IPU2 A0A154PNL8 A0A3L8DGL5 A0A2S2QGV8 A0A1D2MQR4 A0A182P5M9 B4JY03 A0A2S2R6K5 A0A0L0BL37 A0A084VRN6 A0A026WCR2 A0A1A9V4Y0 B4LVX9 E9IGR1 A0A1S4FYN1 A0A2S2P3P2 E2AXJ6 E2C5I1 A0A1A9X419 T1JBE5 A0A1J1J836 Q16JW0 A0A0Q9XEW3 B4K9I3 A0A1A9XT99 A0A1B0BEZ8 A0A3B0JET9 A0A226E6K3 A0A0R1E666 A0A0B4KI64 B4PMJ5 A0A0R3NK67 Q29CA4 B4GNE2 A0A1S5QNT4 A0A067RDH5 B3MT97 A0A2S2NWQ7 A0A1A9ZZN3 B4NGX9 A0A1W4W3S6 A0A1W4VQS7 A0A195C706 A0A151X4Q3 U4UVK4 N6T8W7 A0A0M4EGR7 A0A195D7Y1 A0A158NDV9 F4W7W0 A0A195BQQ4 A0A195ES98 T1I9E8 A0A0P5GI85 A0A0N8DCT7 E9FSA0
A0A0L7KSF3 A0A2A4JRH4 A0A2A3EBV4 A0A087ZTQ6 A0A0L7RBK6 K7IPW7 A0A1B0DQG4 A0A0M8ZTI9 E0VBM5 A0A182UNK7 A0A182HGR8 A0A182JX26 A0A182KMA2 A7UVJ6 W5JDL5 A0A182TGL6 A0A182VQS9 A0A182XVA0 A0A182F6V7 A0A1B0CAC9 W8BAZ6 A0A182RP05 A0A182MTY9 A0A182X2P8 D6W7B0 A0A182N503 A0A182GQN1 A0A0J7NKS2 A0A336MLQ2 A0A336LD69 A0A0C9RHE3 B0W961 A0A1I8N3B3 A0A0K8W735 A0A1I8P0G1 A0A1I8P0A2 A0A1B0FRI8 A0A182IPU2 A0A154PNL8 A0A3L8DGL5 A0A2S2QGV8 A0A1D2MQR4 A0A182P5M9 B4JY03 A0A2S2R6K5 A0A0L0BL37 A0A084VRN6 A0A026WCR2 A0A1A9V4Y0 B4LVX9 E9IGR1 A0A1S4FYN1 A0A2S2P3P2 E2AXJ6 E2C5I1 A0A1A9X419 T1JBE5 A0A1J1J836 Q16JW0 A0A0Q9XEW3 B4K9I3 A0A1A9XT99 A0A1B0BEZ8 A0A3B0JET9 A0A226E6K3 A0A0R1E666 A0A0B4KI64 B4PMJ5 A0A0R3NK67 Q29CA4 B4GNE2 A0A1S5QNT4 A0A067RDH5 B3MT97 A0A2S2NWQ7 A0A1A9ZZN3 B4NGX9 A0A1W4W3S6 A0A1W4VQS7 A0A195C706 A0A151X4Q3 U4UVK4 N6T8W7 A0A0M4EGR7 A0A195D7Y1 A0A158NDV9 F4W7W0 A0A195BQQ4 A0A195ES98 T1I9E8 A0A0P5GI85 A0A0N8DCT7 E9FSA0
Pubmed
19121390
22118469
26354079
26227816
20075255
20566863
+ More
20966253 12364791 14747013 17210077 20920257 23761445 25244985 24495485 18362917 19820115 26483478 25315136 30249741 27289101 17994087 26108605 24438588 24508170 21282665 17510324 20798317 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 23185243 24845553 23537049 21347285 21719571 21292972
20966253 12364791 14747013 17210077 20920257 23761445 25244985 24495485 18362917 19820115 26483478 25315136 30249741 27289101 17994087 26108605 24438588 24508170 21282665 17510324 20798317 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 23185243 24845553 23537049 21347285 21719571 21292972
EMBL
BABH01010312
ODYU01001971
SOQ38905.1
AGBW02008931
OWR52149.1
KQ460779
+ More
KPJ12239.1 KQ459598 KPI94623.1 JTDY01001388 KOB73999.1 JTDY01006240 KOB66177.1 NWSH01000819 PCG74063.1 KZ288291 PBC29217.1 KQ414617 KOC68195.1 AJVK01019108 KQ435851 KOX70840.1 DS235033 EEB10781.1 APCN01005942 APCN01005943 AAAB01008987 EDO63226.2 ADMH02001524 ETN62191.1 AJWK01003658 GAMC01012377 JAB94178.1 AXCM01001504 KQ971307 EFA11053.2 JXUM01080903 KQ563215 KXJ74270.1 LBMM01003849 KMQ93055.1 UFQT01001311 SSX29959.1 UFQS01002871 UFQT01002871 SSX14811.1 SSX34201.1 GBYB01007705 JAG77472.1 DS231862 EDS39758.1 GDHF01005306 JAI47008.1 CCAG010010739 CCAG010010740 KQ434977 KZC12820.1 QOIP01000008 RLU19590.1 GGMS01007774 MBY76977.1 LJIJ01000711 ODM95105.1 CH916377 EDV90565.1 GGMS01016463 MBY85666.1 JRES01001702 KNC20781.1 ATLV01015761 KE525036 KFB40630.1 KK107274 EZA53723.1 CH940650 EDW67584.1 GL763054 EFZ20191.1 GGMR01011474 MBY24093.1 GL443590 EFN61834.1 GL452770 EFN76794.1 JH432010 CVRI01000075 CRL08551.1 CH477989 EAT34582.1 CH933806 KRG02248.1 EDW16643.1 JXJN01013194 OUUW01000005 SPP80894.1 LNIX01000006 OXA52511.1 CM000160 KRK04685.1 AE014297 AGB96497.1 EDW99129.1 CM000070 KRS99522.1 KRS99523.1 EAL26742.2 CH479186 EDW39268.1 KR068532 AMO02549.1 KK852708 KDR17999.1 CH902623 EDV30487.2 GGMR01009052 MBY21671.1 CH964272 EDW84476.1 KQ978251 KYM95948.1 KQ982548 KYQ55269.1 KB632384 ERL94281.1 APGK01047133 KB741077 ENN74148.1 CP012526 ALC47616.1 KQ981153 KYN08962.1 ADTU01013038 ADTU01013039 GL887888 EGI69630.1 KQ976424 KYM88468.1 KQ981993 KYN30777.1 ACPB03002805 GDIQ01240229 JAK11496.1 GDIP01046351 JAM57364.1 GL732523 EFX90357.1
KPJ12239.1 KQ459598 KPI94623.1 JTDY01001388 KOB73999.1 JTDY01006240 KOB66177.1 NWSH01000819 PCG74063.1 KZ288291 PBC29217.1 KQ414617 KOC68195.1 AJVK01019108 KQ435851 KOX70840.1 DS235033 EEB10781.1 APCN01005942 APCN01005943 AAAB01008987 EDO63226.2 ADMH02001524 ETN62191.1 AJWK01003658 GAMC01012377 JAB94178.1 AXCM01001504 KQ971307 EFA11053.2 JXUM01080903 KQ563215 KXJ74270.1 LBMM01003849 KMQ93055.1 UFQT01001311 SSX29959.1 UFQS01002871 UFQT01002871 SSX14811.1 SSX34201.1 GBYB01007705 JAG77472.1 DS231862 EDS39758.1 GDHF01005306 JAI47008.1 CCAG010010739 CCAG010010740 KQ434977 KZC12820.1 QOIP01000008 RLU19590.1 GGMS01007774 MBY76977.1 LJIJ01000711 ODM95105.1 CH916377 EDV90565.1 GGMS01016463 MBY85666.1 JRES01001702 KNC20781.1 ATLV01015761 KE525036 KFB40630.1 KK107274 EZA53723.1 CH940650 EDW67584.1 GL763054 EFZ20191.1 GGMR01011474 MBY24093.1 GL443590 EFN61834.1 GL452770 EFN76794.1 JH432010 CVRI01000075 CRL08551.1 CH477989 EAT34582.1 CH933806 KRG02248.1 EDW16643.1 JXJN01013194 OUUW01000005 SPP80894.1 LNIX01000006 OXA52511.1 CM000160 KRK04685.1 AE014297 AGB96497.1 EDW99129.1 CM000070 KRS99522.1 KRS99523.1 EAL26742.2 CH479186 EDW39268.1 KR068532 AMO02549.1 KK852708 KDR17999.1 CH902623 EDV30487.2 GGMR01009052 MBY21671.1 CH964272 EDW84476.1 KQ978251 KYM95948.1 KQ982548 KYQ55269.1 KB632384 ERL94281.1 APGK01047133 KB741077 ENN74148.1 CP012526 ALC47616.1 KQ981153 KYN08962.1 ADTU01013038 ADTU01013039 GL887888 EGI69630.1 KQ976424 KYM88468.1 KQ981993 KYN30777.1 ACPB03002805 GDIQ01240229 JAK11496.1 GDIP01046351 JAM57364.1 GL732523 EFX90357.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000037510
UP000218220
+ More
UP000242457 UP000005203 UP000053825 UP000002358 UP000092462 UP000053105 UP000009046 UP000075903 UP000075840 UP000075881 UP000075882 UP000007062 UP000000673 UP000075902 UP000075920 UP000076408 UP000069272 UP000092461 UP000075900 UP000075883 UP000076407 UP000007266 UP000075884 UP000069940 UP000249989 UP000036403 UP000002320 UP000095301 UP000095300 UP000092444 UP000075880 UP000076502 UP000279307 UP000094527 UP000075885 UP000001070 UP000037069 UP000030765 UP000053097 UP000078200 UP000008792 UP000000311 UP000008237 UP000091820 UP000183832 UP000008820 UP000009192 UP000092443 UP000092460 UP000268350 UP000198287 UP000002282 UP000000803 UP000001819 UP000008744 UP000027135 UP000007801 UP000092445 UP000007798 UP000192221 UP000078542 UP000075809 UP000030742 UP000019118 UP000092553 UP000078492 UP000005205 UP000007755 UP000078540 UP000078541 UP000015103 UP000000305
UP000242457 UP000005203 UP000053825 UP000002358 UP000092462 UP000053105 UP000009046 UP000075903 UP000075840 UP000075881 UP000075882 UP000007062 UP000000673 UP000075902 UP000075920 UP000076408 UP000069272 UP000092461 UP000075900 UP000075883 UP000076407 UP000007266 UP000075884 UP000069940 UP000249989 UP000036403 UP000002320 UP000095301 UP000095300 UP000092444 UP000075880 UP000076502 UP000279307 UP000094527 UP000075885 UP000001070 UP000037069 UP000030765 UP000053097 UP000078200 UP000008792 UP000000311 UP000008237 UP000091820 UP000183832 UP000008820 UP000009192 UP000092443 UP000092460 UP000268350 UP000198287 UP000002282 UP000000803 UP000001819 UP000008744 UP000027135 UP000007801 UP000092445 UP000007798 UP000192221 UP000078542 UP000075809 UP000030742 UP000019118 UP000092553 UP000078492 UP000005205 UP000007755 UP000078540 UP000078541 UP000015103 UP000000305
PRIDE
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
H9J0K0
A0A2H1VDJ8
A0A212FEH3
A0A194R3Y8
A0A194PNR1
A0A0L7LF39
+ More
A0A0L7KSF3 A0A2A4JRH4 A0A2A3EBV4 A0A087ZTQ6 A0A0L7RBK6 K7IPW7 A0A1B0DQG4 A0A0M8ZTI9 E0VBM5 A0A182UNK7 A0A182HGR8 A0A182JX26 A0A182KMA2 A7UVJ6 W5JDL5 A0A182TGL6 A0A182VQS9 A0A182XVA0 A0A182F6V7 A0A1B0CAC9 W8BAZ6 A0A182RP05 A0A182MTY9 A0A182X2P8 D6W7B0 A0A182N503 A0A182GQN1 A0A0J7NKS2 A0A336MLQ2 A0A336LD69 A0A0C9RHE3 B0W961 A0A1I8N3B3 A0A0K8W735 A0A1I8P0G1 A0A1I8P0A2 A0A1B0FRI8 A0A182IPU2 A0A154PNL8 A0A3L8DGL5 A0A2S2QGV8 A0A1D2MQR4 A0A182P5M9 B4JY03 A0A2S2R6K5 A0A0L0BL37 A0A084VRN6 A0A026WCR2 A0A1A9V4Y0 B4LVX9 E9IGR1 A0A1S4FYN1 A0A2S2P3P2 E2AXJ6 E2C5I1 A0A1A9X419 T1JBE5 A0A1J1J836 Q16JW0 A0A0Q9XEW3 B4K9I3 A0A1A9XT99 A0A1B0BEZ8 A0A3B0JET9 A0A226E6K3 A0A0R1E666 A0A0B4KI64 B4PMJ5 A0A0R3NK67 Q29CA4 B4GNE2 A0A1S5QNT4 A0A067RDH5 B3MT97 A0A2S2NWQ7 A0A1A9ZZN3 B4NGX9 A0A1W4W3S6 A0A1W4VQS7 A0A195C706 A0A151X4Q3 U4UVK4 N6T8W7 A0A0M4EGR7 A0A195D7Y1 A0A158NDV9 F4W7W0 A0A195BQQ4 A0A195ES98 T1I9E8 A0A0P5GI85 A0A0N8DCT7 E9FSA0
A0A0L7KSF3 A0A2A4JRH4 A0A2A3EBV4 A0A087ZTQ6 A0A0L7RBK6 K7IPW7 A0A1B0DQG4 A0A0M8ZTI9 E0VBM5 A0A182UNK7 A0A182HGR8 A0A182JX26 A0A182KMA2 A7UVJ6 W5JDL5 A0A182TGL6 A0A182VQS9 A0A182XVA0 A0A182F6V7 A0A1B0CAC9 W8BAZ6 A0A182RP05 A0A182MTY9 A0A182X2P8 D6W7B0 A0A182N503 A0A182GQN1 A0A0J7NKS2 A0A336MLQ2 A0A336LD69 A0A0C9RHE3 B0W961 A0A1I8N3B3 A0A0K8W735 A0A1I8P0G1 A0A1I8P0A2 A0A1B0FRI8 A0A182IPU2 A0A154PNL8 A0A3L8DGL5 A0A2S2QGV8 A0A1D2MQR4 A0A182P5M9 B4JY03 A0A2S2R6K5 A0A0L0BL37 A0A084VRN6 A0A026WCR2 A0A1A9V4Y0 B4LVX9 E9IGR1 A0A1S4FYN1 A0A2S2P3P2 E2AXJ6 E2C5I1 A0A1A9X419 T1JBE5 A0A1J1J836 Q16JW0 A0A0Q9XEW3 B4K9I3 A0A1A9XT99 A0A1B0BEZ8 A0A3B0JET9 A0A226E6K3 A0A0R1E666 A0A0B4KI64 B4PMJ5 A0A0R3NK67 Q29CA4 B4GNE2 A0A1S5QNT4 A0A067RDH5 B3MT97 A0A2S2NWQ7 A0A1A9ZZN3 B4NGX9 A0A1W4W3S6 A0A1W4VQS7 A0A195C706 A0A151X4Q3 U4UVK4 N6T8W7 A0A0M4EGR7 A0A195D7Y1 A0A158NDV9 F4W7W0 A0A195BQQ4 A0A195ES98 T1I9E8 A0A0P5GI85 A0A0N8DCT7 E9FSA0
PDB
4B6Z
E-value=3.9319e-05,
Score=107
Ontologies
Topology
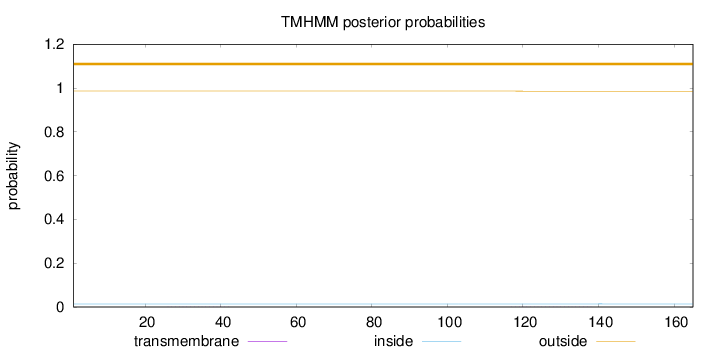
Length:
165
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01508
Exp number, first 60 AAs:
0.00263
Total prob of N-in:
0.01342
outside
1 - 165
Population Genetic Test Statistics
Pi
126.8287
Theta
158.671012
Tajima's D
-0.631031
CLR
1.433112
CSRT
0.209439528023599
Interpretation
Uncertain
