Gene
KWMTBOMO02207
Pre Gene Modal
BGIBMGA003036
Annotation
PREDICTED:_probable_G-protein_coupled_receptor_158_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.881
Sequence
CDS
ATGAGGGCGCGGTGGGCGGCGTGGGCCGTGGCGTGGTGCGCGTGCACCGCCGCCGCGCCATTCGAGCCGCGCGCCTCGCACTCCTCCACTATGCCCTCACCTGTCTGCGATCCACTGCTACTGCACAATCCACCTCCTTTGCCCCTGAATGCACGAGAGCACGAAGCTGCCGTTGTTCACACAGCGCGAGCTCTTGCTGATTTGGTAGAGAAGGGGATAGCAGACGAGGCGCGTGCGACTGCCTTAGCCCGCGCTGTCGCACGCGCTCCGCTCTCGCCGATGCCCGCTGCTGTCGCGCTTGGAGCCCCTCAACTGCATCTCGGAGTACTATACGTCTCGGAACCTTCTCGTCTTATAATATTTCAACAGAACAATTCAGCTACATTACGACAATTCTGGCGTACATTGGCTTCTGCTTGGAATGCAACCAGTGTTGTATGGTCGAACGCTTGGTTTTCTTGTGAAAAAGACGAAGAAGGTTGGTGGGGTGGGATTGCTGCGGCACGAGGCACTGGTCCCACACGTGCAGCAGCTGCAATATTTCGACGTTGGTCGTCACTGCCATGTGAAGAGGCAATGCACGATTGGTTAGGTGAATTTCCGCGATGTGATCCCGTCACTACCGAGTGTGAGGCGTCCAGGGCTCTAAGAGACGGTGAAAGAAAATTAGAGTACAACTGTAAATGTCGACCAGGACATTGGAGTCAGTATGGTGGTGGATGGGTAAACGGCGACTCTTTGGAGCGAAATAATAGGAATCCTTTAACCTGTACCCCATGCGGAATGGGAAGTAGCGCCACCGCTTGTCTTGCTGATGTCTATCGCGTAGGACTACTGGCAGCTAACTTAGCTGGAATGCTATTGTGTGTTGTCATCGGTCTTATTATATTCAGGAAGAGAAAATGCAAGGCGGTTGCCATGGGAATGTGGACTGTATTGGAAGTTGTGTTATTCGGCGCTCTATTAATGTACGCCTCTGCTGCGTCACAGTACATTTCGGTGCCGAAATGGAGGTGTATAGCGGGTGCCTGGCTGCGGGAGCTCGGATTCGTGGCCTGCTATGGATCCGTTGTTTTACGATTATGGGTTTTACTGGCAGAATTTCGTACCCGGAAAGCACACAGGTGGACGCCAAAAGATTCAGAGGTGCTTCGCGTAGTTGGCGCGATGGTACTTGGTGCCGGGTGTTATCTTGCAGCATTCACAGCAACAGCGGCCTGTGAAGAAGACATCGGTGGTTGTGCTAGAGCAGCATGGCACCACGTTACCGGCGCTGCTGAAATCCTACTGCTTCTCGCCGGTATTAGGATCGCTTACGCTGCTAGAAACGCCAATGTTCCGTTTCAGGAGCGTGGGTTGTTGTCAGCGGCGCTGTGGGCGGAGGCGGCTTGCGGCGTGTGGTGGCGTGGCGTGGCGTGGGTCGCCAACGACGCGGCCCCAGAGCGCTGGCCCGTGGCTGCCGTAGCGCGAGCACATCTCTCGTCAGGGGCTGCGCTCGCCTGTGTGCTGGCCCCACGGCTCTGGCACGGCTATCGAGCCAGATCATTAGCTCAAGAGGTTTCTCGTCGAGGGCCCGCAGAAGGTTTTGGCGCAGAAGGTGAAGAAGAGCCCACTTCAGAAGAAGTAAGAGCGGAATTAAAAAGGCTGTACGTCCAACTGGAGATCCTTCGTAACAAGACAATCAGAAGAGATAATCCTCACATCAGTAAAAGACGAGGAGGTCGGAAACCGCCTCATAGACGATTTTCATTACAGGCTCTGCAAGCTCGTCGCGGTGGGAAGGGCAAGCCCAGTGGTGGCGCCAGCGTGGTGGAAGCGAGCGAAGCAGGGGATGCATCGCGGACTCCCGAGGACTCTGTCTGCTCGGTGGAGGGACCCTCACACTACACCGATGGCGACACGCAGGCGTGA
Protein
MRARWAAWAVAWCACTAAAPFEPRASHSSTMPSPVCDPLLLHNPPPLPLNAREHEAAVVHTARALADLVEKGIADEARATALARAVARAPLSPMPAAVALGAPQLHLGVLYVSEPSRLIIFQQNNSATLRQFWRTLASAWNATSVVWSNAWFSCEKDEEGWWGGIAAARGTGPTRAAAAIFRRWSSLPCEEAMHDWLGEFPRCDPVTTECEASRALRDGERKLEYNCKCRPGHWSQYGGGWVNGDSLERNNRNPLTCTPCGMGSSATACLADVYRVGLLAANLAGMLLCVVIGLIIFRKRKCKAVAMGMWTVLEVVLFGALLMYASAASQYISVPKWRCIAGAWLRELGFVACYGSVVLRLWVLLAEFRTRKAHRWTPKDSEVLRVVGAMVLGAGCYLAAFTATAACEEDIGGCARAAWHHVTGAAEILLLLAGIRIAYAARNANVPFQERGLLSAALWAEAACGVWWRGVAWVANDAAPERWPVAAVARAHLSSGAALACVLAPRLWHGYRARSLAQEVSRRGPAEGFGAEGEEEPTSEEVRAELKRLYVQLEILRNKTIRRDNPHISKRRGGRKPPHRRFSLQALQARRGGKGKPSGGASVVEASEAGDASRTPEDSVCSVEGPSHYTDGDTQA
Summary
Uniprot
H9J0J9
A0A2H1VDI7
A0A212FEK8
A0A194R8P2
A0A194PMG3
A0A1W4UX90
+ More
B4I176 A0A0J9QVU7 A0A0Q5W9H0 B4Q339 A0A1W4UVL3 B4NZ22 Q59E18 A0A0J9QVV0 M9PEK9 A0A0Q5WL35 A0A0R1DJ48 B3MVJ3 A0A1I8PH16 A0A0P8XHP9 A0A1I8PGT5 A0A1I8MTN7 A0A1W4UIL0 B3N4B0 A0A0J9QVS7 A0A1W4UI22 M9ND25 A0A0R1DJ99 A0A0J9QVT2 A0A0Q5W9C0 A0A0Q9WXH0 A0A0R1DQS8 A0A0P8XHL4 A0A1I8PH13 A0A0P9A7M0 A0A1I8MTM9 A0A0Q9WLB1 A0A1I8PGT7 B4JQA7 A0A182YMP8 B4G9J6 B4MDQ3 A0A3B0K2B0 B4N0J6 B4KGQ6 A0A1B0APU5 A0A0Q9WLU3 Q29M96 A0A0Q9XF13 A0A0M4E6Y7 A0A1A9WMB5 A0A0M5IXT2 A0A0Q9X620 A0A1B0AGW5 W5JW14 A0A0Q9XFK5 A0A1A9YEY4 A0A1J1IMI7 A0A336MEB0 M9NDF0 A0A1Y1L092 A0A1J1IKL5 A0A336M9V9 A0A1A9VXS8 A0A1B6M2H6 D6WL59 A0A1B6CQI0 A0A139WGZ2 A0A1B0FY78 A0A1B6EH02 A0A1W4UVW1 A0A0Q5WIH6 A0A0J9QWQ0 A0A0R1DJD3 Q9VR40 A0A1I8MTN5 A0A0K8VX51 A0A1I8PH40 A0A0L0CFU0 A0A0A1WMC6 T1IIA3 W8ASF5 A0A182Q291
B4I176 A0A0J9QVU7 A0A0Q5W9H0 B4Q339 A0A1W4UVL3 B4NZ22 Q59E18 A0A0J9QVV0 M9PEK9 A0A0Q5WL35 A0A0R1DJ48 B3MVJ3 A0A1I8PH16 A0A0P8XHP9 A0A1I8PGT5 A0A1I8MTN7 A0A1W4UIL0 B3N4B0 A0A0J9QVS7 A0A1W4UI22 M9ND25 A0A0R1DJ99 A0A0J9QVT2 A0A0Q5W9C0 A0A0Q9WXH0 A0A0R1DQS8 A0A0P8XHL4 A0A1I8PH13 A0A0P9A7M0 A0A1I8MTM9 A0A0Q9WLB1 A0A1I8PGT7 B4JQA7 A0A182YMP8 B4G9J6 B4MDQ3 A0A3B0K2B0 B4N0J6 B4KGQ6 A0A1B0APU5 A0A0Q9WLU3 Q29M96 A0A0Q9XF13 A0A0M4E6Y7 A0A1A9WMB5 A0A0M5IXT2 A0A0Q9X620 A0A1B0AGW5 W5JW14 A0A0Q9XFK5 A0A1A9YEY4 A0A1J1IMI7 A0A336MEB0 M9NDF0 A0A1Y1L092 A0A1J1IKL5 A0A336M9V9 A0A1A9VXS8 A0A1B6M2H6 D6WL59 A0A1B6CQI0 A0A139WGZ2 A0A1B0FY78 A0A1B6EH02 A0A1W4UVW1 A0A0Q5WIH6 A0A0J9QWQ0 A0A0R1DJD3 Q9VR40 A0A1I8MTN5 A0A0K8VX51 A0A1I8PH40 A0A0L0CFU0 A0A0A1WMC6 T1IIA3 W8ASF5 A0A182Q291
Pubmed
EMBL
BABH01010313
ODYU01001971
SOQ38903.1
AGBW02008931
OWR52150.1
KQ460779
+ More
KPJ12241.1 KQ459598 KPI94626.1 CH480820 EDW54283.1 CM002910 KMY88141.1 CH954177 KQS70124.1 CM000361 EDX03755.1 CM000157 EDW87686.1 AE014134 AAX52647.1 KMY88146.1 AGB92592.1 KQS70127.1 KRJ97307.1 CH902624 EDV33258.1 KPU74369.1 EDV57780.2 KMY88142.1 KMY88143.1 KMY88145.1 KMY88147.1 AFH03557.1 KRJ97308.1 KMY88148.1 KQS70125.1 CH940661 KRF85659.1 KRJ97305.1 KPU74368.1 KPU74371.1 KPU74370.1 KRF85660.1 CH916372 EDV99087.1 CH479180 EDW29026.1 EDW71314.2 OUUW01000004 SPP79141.1 CH963920 EDW77609.2 CH933807 EDW13256.1 JXJN01001583 KRF85658.1 CH379060 EAL33798.3 KRG03668.1 CP012523 ALC38427.1 ALC37936.1 KRG03663.1 KRG03664.1 KRG03665.1 KRG03667.1 ADMH02000283 ETN67124.1 KRG03666.1 CVRI01000054 CRL00772.1 UFQT01000775 SSX27133.1 AFH03558.1 GEZM01073528 JAV65026.1 CRL00771.1 SSX27132.1 GEBQ01009836 JAT30141.1 KQ971343 EFA03503.2 GEDC01021627 JAS15671.1 KYB27176.1 AJVK01000779 AJVK01000780 AJVK01000781 AJVK01000782 AJVK01000783 GEDC01000144 JAS37154.1 KQS70126.1 KMY88144.1 KRJ97306.1 AAF50967.3 GDHF01008857 JAI43457.1 JRES01000536 KNC30354.1 GBXI01014285 JAD00007.1 JH430149 GAMC01017523 JAB89032.1 AXCN02000655 AXCN02000656
KPJ12241.1 KQ459598 KPI94626.1 CH480820 EDW54283.1 CM002910 KMY88141.1 CH954177 KQS70124.1 CM000361 EDX03755.1 CM000157 EDW87686.1 AE014134 AAX52647.1 KMY88146.1 AGB92592.1 KQS70127.1 KRJ97307.1 CH902624 EDV33258.1 KPU74369.1 EDV57780.2 KMY88142.1 KMY88143.1 KMY88145.1 KMY88147.1 AFH03557.1 KRJ97308.1 KMY88148.1 KQS70125.1 CH940661 KRF85659.1 KRJ97305.1 KPU74368.1 KPU74371.1 KPU74370.1 KRF85660.1 CH916372 EDV99087.1 CH479180 EDW29026.1 EDW71314.2 OUUW01000004 SPP79141.1 CH963920 EDW77609.2 CH933807 EDW13256.1 JXJN01001583 KRF85658.1 CH379060 EAL33798.3 KRG03668.1 CP012523 ALC38427.1 ALC37936.1 KRG03663.1 KRG03664.1 KRG03665.1 KRG03667.1 ADMH02000283 ETN67124.1 KRG03666.1 CVRI01000054 CRL00772.1 UFQT01000775 SSX27133.1 AFH03558.1 GEZM01073528 JAV65026.1 CRL00771.1 SSX27132.1 GEBQ01009836 JAT30141.1 KQ971343 EFA03503.2 GEDC01021627 JAS15671.1 KYB27176.1 AJVK01000779 AJVK01000780 AJVK01000781 AJVK01000782 AJVK01000783 GEDC01000144 JAS37154.1 KQS70126.1 KMY88144.1 KRJ97306.1 AAF50967.3 GDHF01008857 JAI43457.1 JRES01000536 KNC30354.1 GBXI01014285 JAD00007.1 JH430149 GAMC01017523 JAB89032.1 AXCN02000655 AXCN02000656
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000192221
UP000001292
+ More
UP000008711 UP000000304 UP000002282 UP000000803 UP000007801 UP000095300 UP000095301 UP000008792 UP000001070 UP000076408 UP000008744 UP000268350 UP000007798 UP000009192 UP000092460 UP000001819 UP000092553 UP000091820 UP000092445 UP000000673 UP000092443 UP000183832 UP000078200 UP000007266 UP000092462 UP000037069 UP000075886
UP000008711 UP000000304 UP000002282 UP000000803 UP000007801 UP000095300 UP000095301 UP000008792 UP000001070 UP000076408 UP000008744 UP000268350 UP000007798 UP000009192 UP000092460 UP000001819 UP000092553 UP000091820 UP000092445 UP000000673 UP000092443 UP000183832 UP000078200 UP000007266 UP000092462 UP000037069 UP000075886
Pfam
PF00003 7tm_3
SUPFAM
SSF57184
SSF57184
ProteinModelPortal
H9J0J9
A0A2H1VDI7
A0A212FEK8
A0A194R8P2
A0A194PMG3
A0A1W4UX90
+ More
B4I176 A0A0J9QVU7 A0A0Q5W9H0 B4Q339 A0A1W4UVL3 B4NZ22 Q59E18 A0A0J9QVV0 M9PEK9 A0A0Q5WL35 A0A0R1DJ48 B3MVJ3 A0A1I8PH16 A0A0P8XHP9 A0A1I8PGT5 A0A1I8MTN7 A0A1W4UIL0 B3N4B0 A0A0J9QVS7 A0A1W4UI22 M9ND25 A0A0R1DJ99 A0A0J9QVT2 A0A0Q5W9C0 A0A0Q9WXH0 A0A0R1DQS8 A0A0P8XHL4 A0A1I8PH13 A0A0P9A7M0 A0A1I8MTM9 A0A0Q9WLB1 A0A1I8PGT7 B4JQA7 A0A182YMP8 B4G9J6 B4MDQ3 A0A3B0K2B0 B4N0J6 B4KGQ6 A0A1B0APU5 A0A0Q9WLU3 Q29M96 A0A0Q9XF13 A0A0M4E6Y7 A0A1A9WMB5 A0A0M5IXT2 A0A0Q9X620 A0A1B0AGW5 W5JW14 A0A0Q9XFK5 A0A1A9YEY4 A0A1J1IMI7 A0A336MEB0 M9NDF0 A0A1Y1L092 A0A1J1IKL5 A0A336M9V9 A0A1A9VXS8 A0A1B6M2H6 D6WL59 A0A1B6CQI0 A0A139WGZ2 A0A1B0FY78 A0A1B6EH02 A0A1W4UVW1 A0A0Q5WIH6 A0A0J9QWQ0 A0A0R1DJD3 Q9VR40 A0A1I8MTN5 A0A0K8VX51 A0A1I8PH40 A0A0L0CFU0 A0A0A1WMC6 T1IIA3 W8ASF5 A0A182Q291
B4I176 A0A0J9QVU7 A0A0Q5W9H0 B4Q339 A0A1W4UVL3 B4NZ22 Q59E18 A0A0J9QVV0 M9PEK9 A0A0Q5WL35 A0A0R1DJ48 B3MVJ3 A0A1I8PH16 A0A0P8XHP9 A0A1I8PGT5 A0A1I8MTN7 A0A1W4UIL0 B3N4B0 A0A0J9QVS7 A0A1W4UI22 M9ND25 A0A0R1DJ99 A0A0J9QVT2 A0A0Q5W9C0 A0A0Q9WXH0 A0A0R1DQS8 A0A0P8XHL4 A0A1I8PH13 A0A0P9A7M0 A0A1I8MTM9 A0A0Q9WLB1 A0A1I8PGT7 B4JQA7 A0A182YMP8 B4G9J6 B4MDQ3 A0A3B0K2B0 B4N0J6 B4KGQ6 A0A1B0APU5 A0A0Q9WLU3 Q29M96 A0A0Q9XF13 A0A0M4E6Y7 A0A1A9WMB5 A0A0M5IXT2 A0A0Q9X620 A0A1B0AGW5 W5JW14 A0A0Q9XFK5 A0A1A9YEY4 A0A1J1IMI7 A0A336MEB0 M9NDF0 A0A1Y1L092 A0A1J1IKL5 A0A336M9V9 A0A1A9VXS8 A0A1B6M2H6 D6WL59 A0A1B6CQI0 A0A139WGZ2 A0A1B0FY78 A0A1B6EH02 A0A1W4UVW1 A0A0Q5WIH6 A0A0J9QWQ0 A0A0R1DJD3 Q9VR40 A0A1I8MTN5 A0A0K8VX51 A0A1I8PH40 A0A0L0CFU0 A0A0A1WMC6 T1IIA3 W8ASF5 A0A182Q291
Ontologies
GO
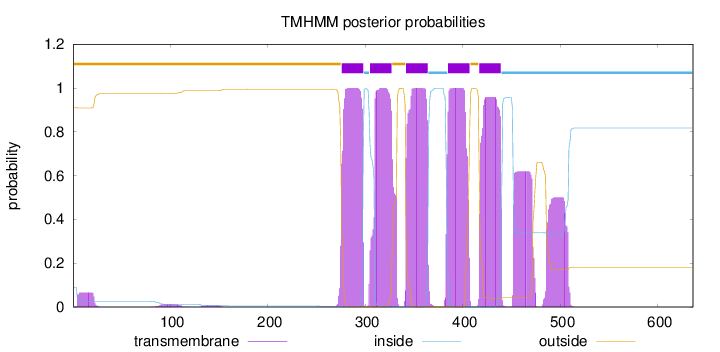
Topology
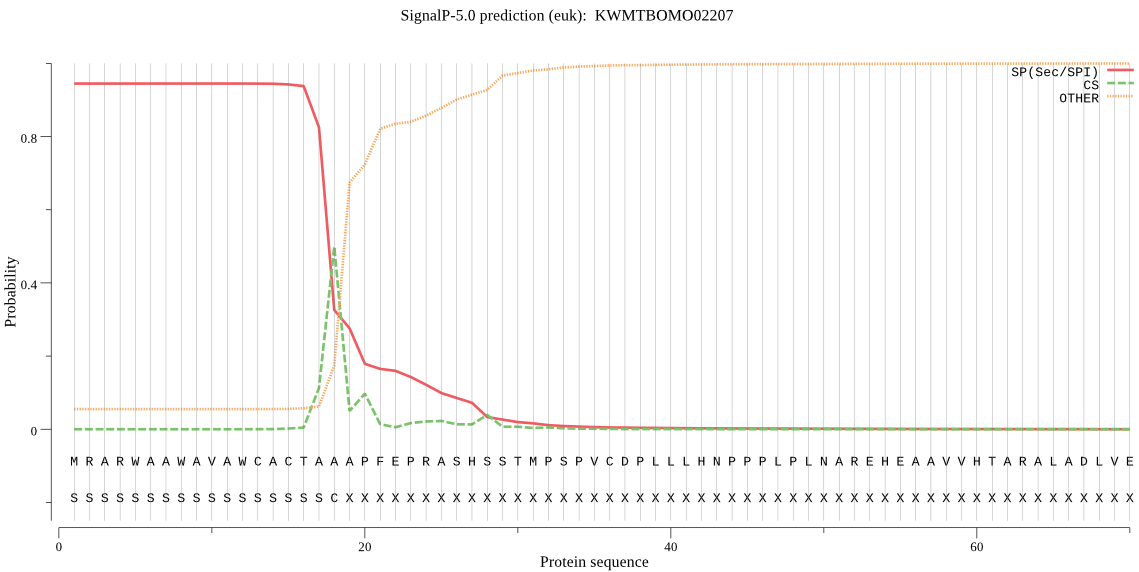
SignalP
Position: 1 - 18,
Likelihood: 0.943967
Length:
636
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
135.25571
Exp number, first 60 AAs:
1.19719
Total prob of N-in:
0.09056
outside
1 - 275
TMhelix
276 - 298
inside
299 - 304
TMhelix
305 - 327
outside
328 - 341
TMhelix
342 - 364
inside
365 - 384
TMhelix
385 - 407
outside
408 - 416
TMhelix
417 - 439
inside
440 - 636
Population Genetic Test Statistics
Pi
36.022828
Theta
23.784058
Tajima's D
-1.009516
CLR
128.234841
CSRT
0.133543322833858
Interpretation
Uncertain
