Pre Gene Modal
BGIBMGA003035
Annotation
PREDICTED:_rab3_GTPase-activating_protein_catalytic_subunit_[Papilio_polytes]
Full name
Rab3 GTPase-activating protein catalytic subunit
Alternative Name
RAB3 GTPase-activating protein 130 kDa subunit
Rab3-GAP p130
Rab3-GAP p130
Location in the cell
Cytoplasmic Reliability : 1.837
Sequence
CDS
ATGAACATCAATGCCAAAAGTCTATGTAATGTCTCTTACAAAGCTCAGAGAATTTTACATGGTGACAGTCACTCTAGCATTGCAGAAGGCACACCCAATGCGCGTACATGCTTACTTCATCAAAAACTACAACTTCTCAACTGCTGCATCAAACGTACCATAGAAGGTAACGTTAACACAACCGGTCATTCTTCTGAGGAAGAGTTCTTCGACTGCTCAGAAGGCGAAGGAGCAGAGGAGCAGCCTCCCTGGGATCGACCTGTCGGCAGACTGCATCGACTCGGTGAAGCCACACTGAAAAATGGCGCGCCGCTATATGTACCACGAACACAAGATCCAGCTCCGAAGACTGAAGATCAATTAGAAGAGGACGCCGAGTTAATGGTGCGCTTGGGAGACGACGCAAAGGCTTCCGAGCTGCGAGCTCGAATGATGAGTGCTTCACTTCTCTCGGATATGGAAGCGTTCAAAGCGGCGAACCCTGGTGCAGAATTATGCGATTTCGTTCGTTGGTATTCCCCAAGAGACTGGAAGTCTGACGATGGAGGTGCCTTAGGTGAACGAATGCTCCTGCCCGGAAATCCTTGGGTTGAAGCTTGGAACGTTGCTCGTCCGGTACCGGCCTCGCGACAGAGACGACTTTTCGATGAAACTCGGGAAGCGGAACAGATTCTACATTTTCTGCGATCCCGCACGGTGAGCGGAGTGGCAGAATTGTTACTTCCCGCGATCATACGAGCAGGAGCGCTGAGAGCTACCCAAGAAGGGGCGATAGGAAGTAGCGCGTCGGTGGGAGGTGTCGACGCGCGCCGAACGTTTGAGATAGCTGTAAGAGAGCTGTATGAGGCTGAGAAAGAAGCTTGCAGAATGCTTTGTCGACGTAAGGTGCTGGGTGGCGGTGAGGAGGGACAGGCGTTGGACGCGGCGGAGAGGGCTCGCGTGCTACGCGCGGCGGGGGGAGTGCTGCCGGCGGTGGCGCGGCGCGAGTTCACGCTGCGGGTGCGGGACGCGACGGCGCCGCAACTCATGCGCGCCGCACTGGCCGACGACATCACCATCGTCGGTGCCTTTACTGAACGTATCGTCTGCTTGTAG
Protein
MNINAKSLCNVSYKAQRILHGDSHSSIAEGTPNARTCLLHQKLQLLNCCIKRTIEGNVNTTGHSSEEEFFDCSEGEGAEEQPPWDRPVGRLHRLGEATLKNGAPLYVPRTQDPAPKTEDQLEEDAELMVRLGDDAKASELRARMMSASLLSDMEAFKAANPGAELCDFVRWYSPRDWKSDDGGALGERMLLPGNPWVEAWNVARPVPASRQRRLFDETREAEQILHFLRSRTVSGVAELLLPAIIRAGALRATQEGAIGSSASVGGVDARRTFEIAVRELYEAEKEACRMLCRRKVLGGGEEGQALDAAERARVLRAAGGVLPAVARREFTLRVRDATAPQLMRAALADDITIVGAFTERIVCL
Summary
Description
Probable catalytic subunit of a GTPase activating protein that has specificity for Rab3 subfamily (RAB3A, RAB3B, RAB3C and RAB3D). Rab3 proteins are involved in regulated exocytosis of neurotransmitters and hormones. Specifically converts active Rab3-GTP to the inactive form Rab3-GDP. Required for normal eye and brain development. May participate in neurodevelopmental processes such as proliferation, migration and differentiation before synapse formation, and non-synaptic vesicular release of neurotransmitters (By similarity).
Subunit
The Rab3 GTPase-activating complex is a heterodimer composed of RAB3GAP and RAB3-GAP150. The Rab3 GTPase-activating complex interacts with DMXL2. Interacts with LMAN1.
The Rab3 GTPase-activating complex is a heterodimer composed of RAB3GAP and RAB3-GAP150. The Rab3 GTPase-activating complex interacts with DMXL2 (PubMed:11809763, PubMed:9733780). Interacts with LMAN1 (By similarity).
The Rab3 GTPase-activating complex is a heterodimer composed of RAB3GAP and RAB3-GAP150. The Rab3 GTPase-activating complex interacts with DMXL2 (PubMed:11809763, PubMed:9733780). Interacts with LMAN1 (By similarity).
Similarity
Belongs to the Rab3-GAP catalytic subunit family.
Keywords
Complete proteome
Cytoplasm
GTPase activation
Phosphoprotein
Reference proteome
Direct protein sequencing
Feature
chain Rab3 GTPase-activating protein catalytic subunit
Uniprot
H9J0J8
A0A212FF94
A0A2A4JQF7
A0A0L7K4D0
A0A2H1VS82
A0A194R4T2
+ More
A0A194PPH4 A0A1Y1KQS1 A0A0L7QM09 Q16WP8 A0A182KGJ8 A0A151I854 Q175P2 A0A1S4G700 A0A069DVL6 A0A154P947 A0A182R3A6 A0A195DZ61 E2BKM0 A0A182MSU1 A0A1D2NF50 A0A182P0I2 A0A182IEL5 A0A088A2F2 A0A1B6DTG6 A0A182TVD3 A0A336M955 A0A182WTH3 A0A182L2C9 A0A182VFU1 A0A182YPN9 A0A182SX11 A0A1S4HDM1 A0A3L8DM64 A0A0A9XWU8 A0A1J1IWU8 A0A3Q0DTC4 A0A1U7T783 L8Y1T2 Q3UYK8 E9HQ35 G1SGN0 Q80UJ7 W5PN04 A0A1D5RLG3 W5PN03 A4FUE5 A0A2I3GP67 L8IEZ5 A0A2Y9SHK8 A0A0G2JXA1 F1LP59 A0A384AYG0 A0A2Y9SKM2 A0A2U4AJG1 A0A384AXU2 A0A2I0MG78 P69735 A0A337SJX5 A0A2Y9MTL8 A0A341D1E4 G1RB30 A0A337SX46 A0A0D9RV02 A0A340XXN4 A0A2Y9N7U1 A0A2Y9N1P5 A0A341CYR2 A0A2U4AJ80 A0A1D5QUM4 A0A0P6K822 A0A2J8T1Z0 A0A2U4AJ87 A0A2U3ZJX9 J9NVN6 A0A2K6AGU1 A0A2K6BW43 G5AMN2
A0A194PPH4 A0A1Y1KQS1 A0A0L7QM09 Q16WP8 A0A182KGJ8 A0A151I854 Q175P2 A0A1S4G700 A0A069DVL6 A0A154P947 A0A182R3A6 A0A195DZ61 E2BKM0 A0A182MSU1 A0A1D2NF50 A0A182P0I2 A0A182IEL5 A0A088A2F2 A0A1B6DTG6 A0A182TVD3 A0A336M955 A0A182WTH3 A0A182L2C9 A0A182VFU1 A0A182YPN9 A0A182SX11 A0A1S4HDM1 A0A3L8DM64 A0A0A9XWU8 A0A1J1IWU8 A0A3Q0DTC4 A0A1U7T783 L8Y1T2 Q3UYK8 E9HQ35 G1SGN0 Q80UJ7 W5PN04 A0A1D5RLG3 W5PN03 A4FUE5 A0A2I3GP67 L8IEZ5 A0A2Y9SHK8 A0A0G2JXA1 F1LP59 A0A384AYG0 A0A2Y9SKM2 A0A2U4AJG1 A0A384AXU2 A0A2I0MG78 P69735 A0A337SJX5 A0A2Y9MTL8 A0A341D1E4 G1RB30 A0A337SX46 A0A0D9RV02 A0A340XXN4 A0A2Y9N7U1 A0A2Y9N1P5 A0A341CYR2 A0A2U4AJ80 A0A1D5QUM4 A0A0P6K822 A0A2J8T1Z0 A0A2U4AJ87 A0A2U3ZJX9 J9NVN6 A0A2K6AGU1 A0A2K6BW43 G5AMN2
Pubmed
19121390
22118469
26227816
26354079
28004739
17510324
+ More
26334808 20798317 27289101 20966253 25244985 30249741 25401762 23385571 10349636 11042159 11076861 11217851 12466851 16141073 21292972 21993624 15368895 16141072 15489334 15696165 21183079 20809919 19468303 19393038 22751099 15057822 23371554 9030515 9733780 11809763 17975172 17431167 16341006 21993625
26334808 20798317 27289101 20966253 25244985 30249741 25401762 23385571 10349636 11042159 11076861 11217851 12466851 16141073 21292972 21993624 15368895 16141072 15489334 15696165 21183079 20809919 19468303 19393038 22751099 15057822 23371554 9030515 9733780 11809763 17975172 17431167 16341006 21993625
EMBL
BABH01010317
BABH01010318
AGBW02008847
OWR52399.1
NWSH01000819
PCG74049.1
+ More
JTDY01011269 KOB57172.1 ODYU01004150 SOQ43713.1 KQ460779 KPJ12250.1 KQ459598 KPI94634.1 GEZM01078342 JAV62761.1 KQ414902 KOC59652.1 CH477557 EAT39019.1 KQ978390 KYM94324.1 CH477397 EAT41808.1 GBGD01000746 JAC88143.1 KQ434847 KZC08466.1 KQ980031 KYN18158.1 GL448819 EFN83718.1 AXCM01006011 LJIJ01000061 ODN03852.1 APCN01005209 GEDC01008335 JAS28963.1 UFQS01000560 UFQT01000560 SSX04961.1 SSX25323.1 QOIP01000006 RLU21497.1 GBHO01021869 GBHO01021868 JAG21735.1 JAG21736.1 CVRI01000063 CRL04576.1 KB368231 ELV10217.1 AK134604 BAE22204.1 GL732714 EFX66131.1 AAGW02005109 AAGW02005110 AAGW02005111 AAGW02005112 AK172881 AK076244 AK076399 AK080432 AK164523 BC028996 BC046297 AMGL01054158 AMGL01054159 AMGL01054160 AMGL01054161 AMGL01054162 AMGL01054163 AMGL01054164 AMGL01054165 AMGL01054166 AMGL01054167 AC163690 BC114755 AAI14756.1 ADFV01039477 ADFV01039478 ADFV01039479 ADFV01039480 ADFV01039481 JH881536 ELR53717.1 AABR07020883 AKCR02000014 PKK28682.1 AABR03085327 AABR03086784 AANG04001482 AQIB01162493 AQIB01162494 AQIB01162495 AQIB01162496 AQIB01162497 JSUE03008637 JSUE03008638 GEBF01000824 JAO02809.1 NDHI03003528 PNJ27047.1 AAEX03011909 AAEX03011910 JH166011 EHA98292.1
JTDY01011269 KOB57172.1 ODYU01004150 SOQ43713.1 KQ460779 KPJ12250.1 KQ459598 KPI94634.1 GEZM01078342 JAV62761.1 KQ414902 KOC59652.1 CH477557 EAT39019.1 KQ978390 KYM94324.1 CH477397 EAT41808.1 GBGD01000746 JAC88143.1 KQ434847 KZC08466.1 KQ980031 KYN18158.1 GL448819 EFN83718.1 AXCM01006011 LJIJ01000061 ODN03852.1 APCN01005209 GEDC01008335 JAS28963.1 UFQS01000560 UFQT01000560 SSX04961.1 SSX25323.1 QOIP01000006 RLU21497.1 GBHO01021869 GBHO01021868 JAG21735.1 JAG21736.1 CVRI01000063 CRL04576.1 KB368231 ELV10217.1 AK134604 BAE22204.1 GL732714 EFX66131.1 AAGW02005109 AAGW02005110 AAGW02005111 AAGW02005112 AK172881 AK076244 AK076399 AK080432 AK164523 BC028996 BC046297 AMGL01054158 AMGL01054159 AMGL01054160 AMGL01054161 AMGL01054162 AMGL01054163 AMGL01054164 AMGL01054165 AMGL01054166 AMGL01054167 AC163690 BC114755 AAI14756.1 ADFV01039477 ADFV01039478 ADFV01039479 ADFV01039480 ADFV01039481 JH881536 ELR53717.1 AABR07020883 AKCR02000014 PKK28682.1 AABR03085327 AABR03086784 AANG04001482 AQIB01162493 AQIB01162494 AQIB01162495 AQIB01162496 AQIB01162497 JSUE03008637 JSUE03008638 GEBF01000824 JAO02809.1 NDHI03003528 PNJ27047.1 AAEX03011909 AAEX03011910 JH166011 EHA98292.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000037510
UP000053240
UP000053268
+ More
UP000053825 UP000008820 UP000075881 UP000078542 UP000076502 UP000075900 UP000078492 UP000008237 UP000075883 UP000094527 UP000075885 UP000075840 UP000005203 UP000075902 UP000076407 UP000075882 UP000075903 UP000076408 UP000075901 UP000279307 UP000183832 UP000189704 UP000011518 UP000000305 UP000001811 UP000000589 UP000002356 UP000009136 UP000001073 UP000248484 UP000002494 UP000261681 UP000245320 UP000053872 UP000011712 UP000248483 UP000252040 UP000029965 UP000265300 UP000006718 UP000245340 UP000002254 UP000233140 UP000233120 UP000006813
UP000053825 UP000008820 UP000075881 UP000078542 UP000076502 UP000075900 UP000078492 UP000008237 UP000075883 UP000094527 UP000075885 UP000075840 UP000005203 UP000075902 UP000076407 UP000075882 UP000075903 UP000076408 UP000075901 UP000279307 UP000183832 UP000189704 UP000011518 UP000000305 UP000001811 UP000000589 UP000002356 UP000009136 UP000001073 UP000248484 UP000002494 UP000261681 UP000245320 UP000053872 UP000011712 UP000248483 UP000252040 UP000029965 UP000265300 UP000006718 UP000245340 UP000002254 UP000233140 UP000233120 UP000006813
PRIDE
Pfam
PF13890 Rab3-GTPase_cat
SUPFAM
SSF47954
SSF47954
CDD
ProteinModelPortal
H9J0J8
A0A212FF94
A0A2A4JQF7
A0A0L7K4D0
A0A2H1VS82
A0A194R4T2
+ More
A0A194PPH4 A0A1Y1KQS1 A0A0L7QM09 Q16WP8 A0A182KGJ8 A0A151I854 Q175P2 A0A1S4G700 A0A069DVL6 A0A154P947 A0A182R3A6 A0A195DZ61 E2BKM0 A0A182MSU1 A0A1D2NF50 A0A182P0I2 A0A182IEL5 A0A088A2F2 A0A1B6DTG6 A0A182TVD3 A0A336M955 A0A182WTH3 A0A182L2C9 A0A182VFU1 A0A182YPN9 A0A182SX11 A0A1S4HDM1 A0A3L8DM64 A0A0A9XWU8 A0A1J1IWU8 A0A3Q0DTC4 A0A1U7T783 L8Y1T2 Q3UYK8 E9HQ35 G1SGN0 Q80UJ7 W5PN04 A0A1D5RLG3 W5PN03 A4FUE5 A0A2I3GP67 L8IEZ5 A0A2Y9SHK8 A0A0G2JXA1 F1LP59 A0A384AYG0 A0A2Y9SKM2 A0A2U4AJG1 A0A384AXU2 A0A2I0MG78 P69735 A0A337SJX5 A0A2Y9MTL8 A0A341D1E4 G1RB30 A0A337SX46 A0A0D9RV02 A0A340XXN4 A0A2Y9N7U1 A0A2Y9N1P5 A0A341CYR2 A0A2U4AJ80 A0A1D5QUM4 A0A0P6K822 A0A2J8T1Z0 A0A2U4AJ87 A0A2U3ZJX9 J9NVN6 A0A2K6AGU1 A0A2K6BW43 G5AMN2
A0A194PPH4 A0A1Y1KQS1 A0A0L7QM09 Q16WP8 A0A182KGJ8 A0A151I854 Q175P2 A0A1S4G700 A0A069DVL6 A0A154P947 A0A182R3A6 A0A195DZ61 E2BKM0 A0A182MSU1 A0A1D2NF50 A0A182P0I2 A0A182IEL5 A0A088A2F2 A0A1B6DTG6 A0A182TVD3 A0A336M955 A0A182WTH3 A0A182L2C9 A0A182VFU1 A0A182YPN9 A0A182SX11 A0A1S4HDM1 A0A3L8DM64 A0A0A9XWU8 A0A1J1IWU8 A0A3Q0DTC4 A0A1U7T783 L8Y1T2 Q3UYK8 E9HQ35 G1SGN0 Q80UJ7 W5PN04 A0A1D5RLG3 W5PN03 A4FUE5 A0A2I3GP67 L8IEZ5 A0A2Y9SHK8 A0A0G2JXA1 F1LP59 A0A384AYG0 A0A2Y9SKM2 A0A2U4AJG1 A0A384AXU2 A0A2I0MG78 P69735 A0A337SJX5 A0A2Y9MTL8 A0A341D1E4 G1RB30 A0A337SX46 A0A0D9RV02 A0A340XXN4 A0A2Y9N7U1 A0A2Y9N1P5 A0A341CYR2 A0A2U4AJ80 A0A1D5QUM4 A0A0P6K822 A0A2J8T1Z0 A0A2U4AJ87 A0A2U3ZJX9 J9NVN6 A0A2K6AGU1 A0A2K6BW43 G5AMN2
Ontologies
GO
GO:0005096
GO:0016021
GO:0043547
GO:0098794
GO:0060325
GO:0017112
GO:0005811
GO:0005794
GO:0061646
GO:0034389
GO:1903061
GO:1903373
GO:0060079
GO:2000786
GO:0097051
GO:0032991
GO:0071782
GO:0010628
GO:1903233
GO:0048172
GO:0043010
GO:0021854
GO:0048489
GO:0017137
GO:0032483
GO:0050821
GO:0043087
GO:0007420
GO:0005737
GO:0010807
GO:0046982
GO:0060076
GO:0005680
GO:0030071
GO:0016020
GO:0005839
GO:0051603
GO:0008483
GO:0006418
GO:0016507
GO:0048384
PANTHER
Topology
Subcellular location
Cytoplasm
In neurons, it is enriched in the synaptic soluble fraction. With evidence from 2 publications.
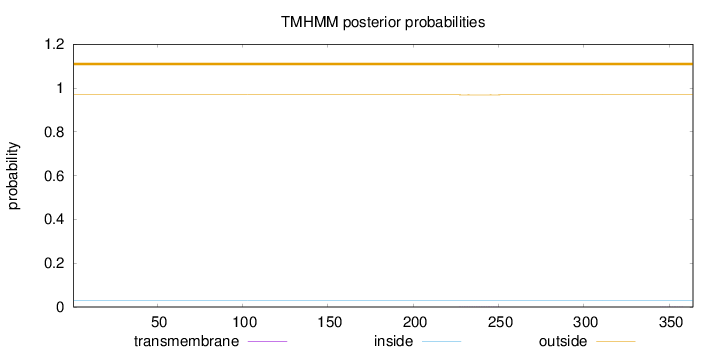
Length:
364
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01634
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03117
outside
1 - 364
Population Genetic Test Statistics
Pi
241.509104
Theta
175.699404
Tajima's D
0.626423
CLR
0.016657
CSRT
0.556922153892305
Interpretation
Uncertain
