Gene
KWMTBOMO02205
Pre Gene Modal
BGIBMGA003034
Annotation
Uncharacterized_protein_OBRU01_12362?_partial_[Operophtera_brumata]
Full name
Rab3 GTPase-activating protein catalytic subunit
Alternative Name
RAB3 GTPase-activating protein 130 kDa subunit
Rab3-GAP p130
Rab3-GAP p130
Location in the cell
Cytoplasmic Reliability : 1.683
Sequence
CDS
ATGAATGAAGAAATAGATGAATACGACATTTACCACCAAGATTTTACTACCGTATCAGAATGGGAGGTATTTGTAGCACGAATAGAAGAGGTACTGATTGATTGGAGACTGTCTAAATCCACAATGGTTAAGCATTCTGTTGAAAATCATGCAAAAAAATGGTTTACAAAAAGCGAAGAAATAAGCTATCAAGGACAGAAATTTACTCTTTCTTATAATTATATGAACTGTAAAAGTAAACCAGAAAGCACTGAAGAATCAAGTGATAAATCATTGTTGGTCGAAATCCAAGATGGACTTTGGGAAGCCACGACTAGGTTTATGGATGACAGTGACGAGAGTCCCTTTCCTGTGTCAAACTGGTTTGGTTTAAAACAATATTTAATGTTGTATCCTAATGTAGCACTTGTTGAAGGTAGTTTAATTAAACTGGTTATGAGTAGTTTAAACATAGCATTTGCAAATGTTGATTGTGGTGTACCTTTATTTGTTAAGATAAGAGAACACTGGCAACATACTTATTTAGGATTCTATGAAGACAATGAATATAAGATAAGCTTTGACACTATTCATTTAAAGAGGATATCCAATCAGTGCTCCCACTTAACTGGCCTCGTTGGATTATTTAAGTCAAAAATTGCTTCACCAGTGGCATTGGATGCTGTTGTTGTTTCAACAAAGTTTTCATATGAATTGAAAGATTTAGATGCTTTTGCATGGGGTCAGCAAACACAGTCAAACGAGTTTTTGTCTATAGATGGTTTTGATGTAAAGAAGTTGATTGAACTGCCATTTGGCTGTGATAAGAACCCCACAAAAAGTGTATTACTTAATGCAATATGGCCAAAATTTAGAGAATGCACAATAGTGGAATCATCTACGTTTTCAGATATCGATCCATTAATGGCACCAACCTGGACTCTAACTTTAAAGATGAGAGAAAATATAAATTTCCAATTGTCAGAATTAATAGGGAAGGTGATGGAACTCTTAGATAATTCTTATCTATTAATGGACACTCTTGGCTTGAGTAAAAGTCTAGGACTAGTTAACCCATTAAACAAGATTACAGAAGCTCCTATTACTATATCCAAATTGGTGAAGGCTGCAATGGGTCATGGTCGTCATACATCTGAATTCAAGGGGCCTATATCAGATGAACTGTTAATGCCCCTTTTATACTATATATTTCCTGATGCTGTTGAAGATGGCACTCATATTTATCCTCCTGATTTGAAAATGGAATTTAAACAGGATGGTAATGGATCTAAGCTATGCAGCTATGTAAAGAGTAGCCCTGAAGATTGCCTCGTCTGGAGATTATCAGTAACTGCAGCGAAACTGCATGATGCTGGTGGTCTTTCATATTTAGCACATTTGTGGTATGAATTCATTCAAGAGCTGCAGTATAGATGGGAGATGCAAATACCAGTTCCTGGGTAA
Protein
MNEEIDEYDIYHQDFTTVSEWEVFVARIEEVLIDWRLSKSTMVKHSVENHAKKWFTKSEEISYQGQKFTLSYNYMNCKSKPESTEESSDKSLLVEIQDGLWEATTRFMDDSDESPFPVSNWFGLKQYLMLYPNVALVEGSLIKLVMSSLNIAFANVDCGVPLFVKIREHWQHTYLGFYEDNEYKISFDTIHLKRISNQCSHLTGLVGLFKSKIASPVALDAVVVSTKFSYELKDLDAFAWGQQTQSNEFLSIDGFDVKKLIELPFGCDKNPTKSVLLNAIWPKFRECTIVESSTFSDIDPLMAPTWTLTLKMRENINFQLSELIGKVMELLDNSYLLMDTLGLSKSLGLVNPLNKITEAPITISKLVKAAMGHGRHTSEFKGPISDELLMPLLYYIFPDAVEDGTHIYPPDLKMEFKQDGNGSKLCSYVKSSPEDCLVWRLSVTAAKLHDAGGLSYLAHLWYEFIQELQYRWEMQIPVPG
Summary
Description
Probable catalytic subunit of a GTPase activating protein that has specificity for Rab3 subfamily. Rab3 proteins are involved in regulated exocytosis of neurotransmitters and hormones. Specifically converts active Rab3-GTP to the inactive form Rab3-GDP (By similarity).
Probable catalytic subunit of a GTPase activating protein that has specificity for Rab3 subfamily (RAB3A, RAB3B, RAB3C and RAB3D). Rab3 proteins are involved in regulated exocytosis of neurotransmitters and hormones. Specifically converts active Rab3-GTP to the inactive form Rab3-GDP. Required for normal eye and brain development. May participate in neurodevelopmental processes such as proliferation, migration and differentiation before synapse formation, and non-synaptic vesicular release of neurotransmitters.
Probable catalytic subunit of a GTPase activating protein that has specificity for Rab3 subfamily (RAB3A, RAB3B, RAB3C and RAB3D). Rab3 proteins are involved in regulated exocytosis of neurotransmitters and hormones. Specifically converts active Rab3-GTP to the inactive form Rab3-GDP. Required for normal eye and brain development. May participate in neurodevelopmental processes such as proliferation, migration and differentiation before synapse formation, and non-synaptic vesicular release of neurotransmitters.
Biophysicochemical Properties
75 uM for GTP-loaded RAB3A
Subunit
The Rab3 GTPase-activating complex is a heterodimer composed of RAB3GAP and RAB3GAP150.
The Rab3 GTPase-activating complex is a heterodimer composed of RAB3GAP and RAB3-GAP150. The Rab3 GTPase-activating complex interacts with DMXL2 (By similarity). Interacts with LMAN1 (PubMed:22337587).
The Rab3 GTPase-activating complex is a heterodimer composed of RAB3GAP and RAB3-GAP150. The Rab3 GTPase-activating complex interacts with DMXL2 (By similarity). Interacts with LMAN1 (PubMed:22337587).
Similarity
Belongs to the Rab3-GAP catalytic subunit family.
Keywords
Cytoplasm
GTPase activation
Alternative splicing
Complete proteome
Phosphoprotein
Polymorphism
Reference proteome
Feature
chain Rab3 GTPase-activating protein catalytic subunit
splice variant In isoform 3.
sequence variant In dbSNP:rs10445686.
splice variant In isoform 3.
sequence variant In dbSNP:rs10445686.
Uniprot
H9J0J7
A0A0L7LAD9
A0A2H1X357
A0A2A4JB34
A0A212FFC8
A0A194PPH4
+ More
A0A194R3I1 A0A1E1W3U9 A0A1E1WB08 A0A1B6FGQ0 D6WL62 A0A0T6BA24 A0A1B6LTE9 A0A1W4XJK2 A0A158NK38 F4WIJ7 A0A1Y1KQS1 E2A107 E0VQA7 A0A195F493 A0A026WD15 A0A0J7NZY6 A0A232EZ14 A0A195DZ61 A0A0L7QM27 E9JCD8 A0A0A9XWU8 A0A088A2F1 A0A2P8XBJ8 A0A310SPL9 N6THU8 A0A2A3E9U2 A0A154P9F7 E2BKM2 A0A3L8DM64 K7JQL9 A0A182F603 A0A182R3A8 A0A182IYR2 A0A182GQC3 A0A1L8EVK4 Q642R9 A0A084W3W1 A0A250YM54 A0A2M4CQ80 A0A151I854 A0A1L8ENU5 A0A2M4ANB3 W5J2M7 A0A182P0I0 A0A2D0R6Z6 G1RB30 R4G9R7 A0A2D0R4U4 A0A0N8A333 Q5RF09 A0A2K5I373 A0A286X9L1 A0A2K5I2U4 H0VCU4 B9A6J2 Q15042 Q15042-3 A0A2J8T1Z3 A0A2Y9HII3 A0A2R9A751 K7BLE5 G3RGG4 A0A2R9A2X7 H2QIR5 F6QUK1 U6DJQ4 H2P7F3 A0A2J8T219 A0A2K6PQY6 W5PN04 A0A2K6MXP7 A0A2K6MXQ1 A0A2K6MXP9 G3SA53 A0A1S3GDG3 A0A2K6PQX1 A0A2U3W640 A0A2Y9L322 A0A3Q0D6G3 A0A2K6PR00 A0A2Y9L9H9 W5M0U5 W5M0S6 M3XTV7 A4FUE5 A0A2Y9HPS2 L8IEZ5 A0A0P5T9Z5 G1MDA5 A0A182MDG4 A0A2U3W615 G1MDA3 K9INK1 A0A2D4FK10
A0A194R3I1 A0A1E1W3U9 A0A1E1WB08 A0A1B6FGQ0 D6WL62 A0A0T6BA24 A0A1B6LTE9 A0A1W4XJK2 A0A158NK38 F4WIJ7 A0A1Y1KQS1 E2A107 E0VQA7 A0A195F493 A0A026WD15 A0A0J7NZY6 A0A232EZ14 A0A195DZ61 A0A0L7QM27 E9JCD8 A0A0A9XWU8 A0A088A2F1 A0A2P8XBJ8 A0A310SPL9 N6THU8 A0A2A3E9U2 A0A154P9F7 E2BKM2 A0A3L8DM64 K7JQL9 A0A182F603 A0A182R3A8 A0A182IYR2 A0A182GQC3 A0A1L8EVK4 Q642R9 A0A084W3W1 A0A250YM54 A0A2M4CQ80 A0A151I854 A0A1L8ENU5 A0A2M4ANB3 W5J2M7 A0A182P0I0 A0A2D0R6Z6 G1RB30 R4G9R7 A0A2D0R4U4 A0A0N8A333 Q5RF09 A0A2K5I373 A0A286X9L1 A0A2K5I2U4 H0VCU4 B9A6J2 Q15042 Q15042-3 A0A2J8T1Z3 A0A2Y9HII3 A0A2R9A751 K7BLE5 G3RGG4 A0A2R9A2X7 H2QIR5 F6QUK1 U6DJQ4 H2P7F3 A0A2J8T219 A0A2K6PQY6 W5PN04 A0A2K6MXP7 A0A2K6MXQ1 A0A2K6MXP9 G3SA53 A0A1S3GDG3 A0A2K6PQX1 A0A2U3W640 A0A2Y9L322 A0A3Q0D6G3 A0A2K6PR00 A0A2Y9L9H9 W5M0U5 W5M0S6 M3XTV7 A4FUE5 A0A2Y9HPS2 L8IEZ5 A0A0P5T9Z5 G1MDA5 A0A182MDG4 A0A2U3W615 G1MDA3 K9INK1 A0A2D4FK10
Pubmed
19121390
26227816
22118469
26354079
18362917
19820115
+ More
21347285 21719571 28004739 20798317 20566863 24508170 28648823 21282665 25401762 29403074 23537049 30249741 20075255 26483478 27762356 24438588 28087693 20920257 23761445 21993624 11181995 19077034 7584044 15815621 15489334 17974005 9030515 9852129 10859313 15696165 17081983 18669648 19690332 20068231 21269460 22337587 23186163 24275569 22722832 16136131 22398555 19892987 25362486 20809919 26319212 19393038 22751099 20010809
21347285 21719571 28004739 20798317 20566863 24508170 28648823 21282665 25401762 29403074 23537049 30249741 20075255 26483478 27762356 24438588 28087693 20920257 23761445 21993624 11181995 19077034 7584044 15815621 15489334 17974005 9030515 9852129 10859313 15696165 17081983 18669648 19690332 20068231 21269460 22337587 23186163 24275569 22722832 16136131 22398555 19892987 25362486 20809919 26319212 19393038 22751099 20010809
EMBL
BABH01010319
JTDY01001995
KOB72375.1
ODYU01013080
SOQ59761.1
NWSH01002294
+ More
PCG68640.1 AGBW02008830 OWR52430.1 KQ459598 KPI94634.1 KQ460779 KPJ12252.1 GDQN01009432 JAT81622.1 GDQN01006929 JAT84125.1 GECZ01020616 JAS49153.1 KQ971343 EFA03501.1 LJIG01002764 KRT84176.1 GEBQ01013033 JAT26944.1 ADTU01018485 GL888176 EGI65933.1 GEZM01078342 JAV62761.1 GL435626 EFN73060.1 DS235408 EEB15563.1 KQ981820 KYN35283.1 KK107282 EZA53556.1 LBMM01000615 KMQ97965.1 NNAY01001561 OXU23595.1 KQ980031 KYN18158.1 KQ414902 KOC59650.1 GL771786 EFZ09513.1 GBHO01021869 GBHO01021868 JAG21735.1 JAG21736.1 PYGN01004048 PSN29377.1 KQ761976 OAD56412.1 APGK01037626 KB740948 KB632364 ENN77388.1 ERL93608.1 KZ288311 PBC28497.1 KQ434847 KZC08463.1 GL448819 EFN83720.1 QOIP01000006 RLU21497.1 JXUM01015628 KQ560435 KXJ82431.1 CM004482 OCT63360.1 BC081089 ATLV01020162 ATLV01020163 ATLV01020164 ATLV01020165 ATLV01020166 ATLV01020167 ATLV01020168 ATLV01020169 ATLV01020170 KE525295 KFB44905.1 GFFW01000202 JAV44586.1 GGFL01003318 MBW67496.1 KQ978390 KYM94324.1 CM004483 OCT60981.1 GGFK01008777 MBW42098.1 ADMH02002182 ETN58006.1 ADFV01039477 ADFV01039478 ADFV01039479 ADFV01039480 ADFV01039481 GDIP01187273 JAJ36129.1 CR857352 CAH89648.1 AAKN02006578 AAKN02006579 AAKN02006580 AAKN02006581 AAKN02006582 AB449877 CH471058 BAH16620.1 EAX11641.1 D31886 AC017031 AC020602 BC022977 BC071602 BC146809 AL096752 NDHI03003528 PNJ27048.1 AJFE02079619 AJFE02079620 AJFE02079621 AACZ04058336 AACZ04058337 GABC01008959 GABF01008915 GABD01002029 GABE01002459 NBAG03000211 JAA02379.1 JAA13230.1 JAA31071.1 JAA42280.1 PNI85722.1 CABD030015916 CABD030015917 CABD030015918 PNI85723.1 HAAF01011002 CCP82826.1 ABGA01194984 ABGA01194985 ABGA01194986 ABGA01194987 ABGA01194988 ABGA01194989 ABGA01194990 ABGA01194991 ABGA01194992 PNJ27049.1 AMGL01054158 AMGL01054159 AMGL01054160 AMGL01054161 AMGL01054162 AMGL01054163 AMGL01054164 AMGL01054165 AMGL01054166 AMGL01054167 AHAT01011639 AEYP01083417 AEYP01083418 AEYP01083419 AEYP01083420 AEYP01083421 AEYP01083422 AEYP01083423 AEYP01083424 BC114755 AAI14756.1 JH881536 ELR53717.1 GDIP01133136 JAL70578.1 ACTA01066452 ACTA01074452 ACTA01082452 ACTA01090452 AXCM01006971 GABZ01004062 JAA49463.1 IACJ01076199 LAA47845.1
PCG68640.1 AGBW02008830 OWR52430.1 KQ459598 KPI94634.1 KQ460779 KPJ12252.1 GDQN01009432 JAT81622.1 GDQN01006929 JAT84125.1 GECZ01020616 JAS49153.1 KQ971343 EFA03501.1 LJIG01002764 KRT84176.1 GEBQ01013033 JAT26944.1 ADTU01018485 GL888176 EGI65933.1 GEZM01078342 JAV62761.1 GL435626 EFN73060.1 DS235408 EEB15563.1 KQ981820 KYN35283.1 KK107282 EZA53556.1 LBMM01000615 KMQ97965.1 NNAY01001561 OXU23595.1 KQ980031 KYN18158.1 KQ414902 KOC59650.1 GL771786 EFZ09513.1 GBHO01021869 GBHO01021868 JAG21735.1 JAG21736.1 PYGN01004048 PSN29377.1 KQ761976 OAD56412.1 APGK01037626 KB740948 KB632364 ENN77388.1 ERL93608.1 KZ288311 PBC28497.1 KQ434847 KZC08463.1 GL448819 EFN83720.1 QOIP01000006 RLU21497.1 JXUM01015628 KQ560435 KXJ82431.1 CM004482 OCT63360.1 BC081089 ATLV01020162 ATLV01020163 ATLV01020164 ATLV01020165 ATLV01020166 ATLV01020167 ATLV01020168 ATLV01020169 ATLV01020170 KE525295 KFB44905.1 GFFW01000202 JAV44586.1 GGFL01003318 MBW67496.1 KQ978390 KYM94324.1 CM004483 OCT60981.1 GGFK01008777 MBW42098.1 ADMH02002182 ETN58006.1 ADFV01039477 ADFV01039478 ADFV01039479 ADFV01039480 ADFV01039481 GDIP01187273 JAJ36129.1 CR857352 CAH89648.1 AAKN02006578 AAKN02006579 AAKN02006580 AAKN02006581 AAKN02006582 AB449877 CH471058 BAH16620.1 EAX11641.1 D31886 AC017031 AC020602 BC022977 BC071602 BC146809 AL096752 NDHI03003528 PNJ27048.1 AJFE02079619 AJFE02079620 AJFE02079621 AACZ04058336 AACZ04058337 GABC01008959 GABF01008915 GABD01002029 GABE01002459 NBAG03000211 JAA02379.1 JAA13230.1 JAA31071.1 JAA42280.1 PNI85722.1 CABD030015916 CABD030015917 CABD030015918 PNI85723.1 HAAF01011002 CCP82826.1 ABGA01194984 ABGA01194985 ABGA01194986 ABGA01194987 ABGA01194988 ABGA01194989 ABGA01194990 ABGA01194991 ABGA01194992 PNJ27049.1 AMGL01054158 AMGL01054159 AMGL01054160 AMGL01054161 AMGL01054162 AMGL01054163 AMGL01054164 AMGL01054165 AMGL01054166 AMGL01054167 AHAT01011639 AEYP01083417 AEYP01083418 AEYP01083419 AEYP01083420 AEYP01083421 AEYP01083422 AEYP01083423 AEYP01083424 BC114755 AAI14756.1 JH881536 ELR53717.1 GDIP01133136 JAL70578.1 ACTA01066452 ACTA01074452 ACTA01082452 ACTA01090452 AXCM01006971 GABZ01004062 JAA49463.1 IACJ01076199 LAA47845.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000007266 UP000192223 UP000005205 UP000007755 UP000000311 UP000009046 UP000078541 UP000053097 UP000036403 UP000215335 UP000078492 UP000053825 UP000005203 UP000245037 UP000019118 UP000030742 UP000242457 UP000076502 UP000008237 UP000279307 UP000002358 UP000069272 UP000075900 UP000075880 UP000069940 UP000249989 UP000186698 UP000030765 UP000078542 UP000000673 UP000075885 UP000221080 UP000001073 UP000001646 UP000233080 UP000005447 UP000005640 UP000248481 UP000240080 UP000002277 UP000001519 UP000002281 UP000001595 UP000233200 UP000002356 UP000233180 UP000081671 UP000245340 UP000248482 UP000189706 UP000018468 UP000000715 UP000009136 UP000008912 UP000075883
UP000007266 UP000192223 UP000005205 UP000007755 UP000000311 UP000009046 UP000078541 UP000053097 UP000036403 UP000215335 UP000078492 UP000053825 UP000005203 UP000245037 UP000019118 UP000030742 UP000242457 UP000076502 UP000008237 UP000279307 UP000002358 UP000069272 UP000075900 UP000075880 UP000069940 UP000249989 UP000186698 UP000030765 UP000078542 UP000000673 UP000075885 UP000221080 UP000001073 UP000001646 UP000233080 UP000005447 UP000005640 UP000248481 UP000240080 UP000002277 UP000001519 UP000002281 UP000001595 UP000233200 UP000002356 UP000233180 UP000081671 UP000245340 UP000248482 UP000189706 UP000018468 UP000000715 UP000009136 UP000008912 UP000075883
Pfam
PF13890 Rab3-GTPase_cat
Interpro
IPR026147
Rab3-GAP_cat_su
ProteinModelPortal
H9J0J7
A0A0L7LAD9
A0A2H1X357
A0A2A4JB34
A0A212FFC8
A0A194PPH4
+ More
A0A194R3I1 A0A1E1W3U9 A0A1E1WB08 A0A1B6FGQ0 D6WL62 A0A0T6BA24 A0A1B6LTE9 A0A1W4XJK2 A0A158NK38 F4WIJ7 A0A1Y1KQS1 E2A107 E0VQA7 A0A195F493 A0A026WD15 A0A0J7NZY6 A0A232EZ14 A0A195DZ61 A0A0L7QM27 E9JCD8 A0A0A9XWU8 A0A088A2F1 A0A2P8XBJ8 A0A310SPL9 N6THU8 A0A2A3E9U2 A0A154P9F7 E2BKM2 A0A3L8DM64 K7JQL9 A0A182F603 A0A182R3A8 A0A182IYR2 A0A182GQC3 A0A1L8EVK4 Q642R9 A0A084W3W1 A0A250YM54 A0A2M4CQ80 A0A151I854 A0A1L8ENU5 A0A2M4ANB3 W5J2M7 A0A182P0I0 A0A2D0R6Z6 G1RB30 R4G9R7 A0A2D0R4U4 A0A0N8A333 Q5RF09 A0A2K5I373 A0A286X9L1 A0A2K5I2U4 H0VCU4 B9A6J2 Q15042 Q15042-3 A0A2J8T1Z3 A0A2Y9HII3 A0A2R9A751 K7BLE5 G3RGG4 A0A2R9A2X7 H2QIR5 F6QUK1 U6DJQ4 H2P7F3 A0A2J8T219 A0A2K6PQY6 W5PN04 A0A2K6MXP7 A0A2K6MXQ1 A0A2K6MXP9 G3SA53 A0A1S3GDG3 A0A2K6PQX1 A0A2U3W640 A0A2Y9L322 A0A3Q0D6G3 A0A2K6PR00 A0A2Y9L9H9 W5M0U5 W5M0S6 M3XTV7 A4FUE5 A0A2Y9HPS2 L8IEZ5 A0A0P5T9Z5 G1MDA5 A0A182MDG4 A0A2U3W615 G1MDA3 K9INK1 A0A2D4FK10
A0A194R3I1 A0A1E1W3U9 A0A1E1WB08 A0A1B6FGQ0 D6WL62 A0A0T6BA24 A0A1B6LTE9 A0A1W4XJK2 A0A158NK38 F4WIJ7 A0A1Y1KQS1 E2A107 E0VQA7 A0A195F493 A0A026WD15 A0A0J7NZY6 A0A232EZ14 A0A195DZ61 A0A0L7QM27 E9JCD8 A0A0A9XWU8 A0A088A2F1 A0A2P8XBJ8 A0A310SPL9 N6THU8 A0A2A3E9U2 A0A154P9F7 E2BKM2 A0A3L8DM64 K7JQL9 A0A182F603 A0A182R3A8 A0A182IYR2 A0A182GQC3 A0A1L8EVK4 Q642R9 A0A084W3W1 A0A250YM54 A0A2M4CQ80 A0A151I854 A0A1L8ENU5 A0A2M4ANB3 W5J2M7 A0A182P0I0 A0A2D0R6Z6 G1RB30 R4G9R7 A0A2D0R4U4 A0A0N8A333 Q5RF09 A0A2K5I373 A0A286X9L1 A0A2K5I2U4 H0VCU4 B9A6J2 Q15042 Q15042-3 A0A2J8T1Z3 A0A2Y9HII3 A0A2R9A751 K7BLE5 G3RGG4 A0A2R9A2X7 H2QIR5 F6QUK1 U6DJQ4 H2P7F3 A0A2J8T219 A0A2K6PQY6 W5PN04 A0A2K6MXP7 A0A2K6MXQ1 A0A2K6MXP9 G3SA53 A0A1S3GDG3 A0A2K6PQX1 A0A2U3W640 A0A2Y9L322 A0A3Q0D6G3 A0A2K6PR00 A0A2Y9L9H9 W5M0U5 W5M0S6 M3XTV7 A4FUE5 A0A2Y9HPS2 L8IEZ5 A0A0P5T9Z5 G1MDA5 A0A182MDG4 A0A2U3W615 G1MDA3 K9INK1 A0A2D4FK10
Ontologies
GO
GO:0005096
GO:0016021
GO:0043547
GO:0005737
GO:0060325
GO:0098794
GO:2000786
GO:0097051
GO:0071782
GO:0032991
GO:0005794
GO:1903233
GO:0010628
GO:0017112
GO:0005811
GO:0061646
GO:0034389
GO:1903373
GO:0048172
GO:1903061
GO:0021854
GO:0043010
GO:0060079
GO:0017137
GO:0070062
GO:0005085
GO:0005789
GO:0007420
GO:0043087
GO:0005829
PANTHER
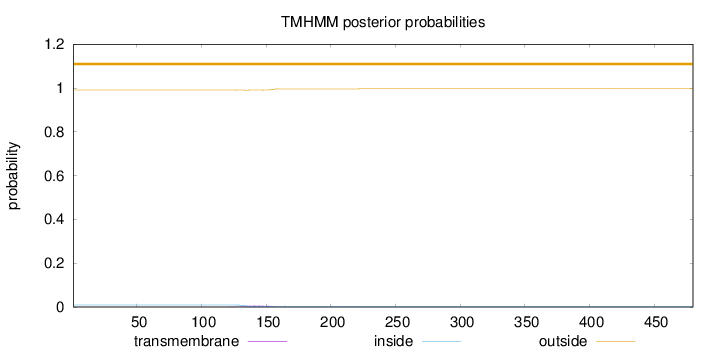
Topology
Subcellular location
Cytoplasm
Length:
480
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14752
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00925
outside
1 - 480
Population Genetic Test Statistics
Pi
231.265041
Theta
201.061511
Tajima's D
0.44482
CLR
1.053044
CSRT
0.494175291235438
Interpretation
Uncertain
