Gene
KWMTBOMO02202
Pre Gene Modal
BGIBMGA003124
Annotation
PREDICTED:_cell_division_cycle_protein_23_homolog_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.38 Nuclear Reliability : 1.345
Sequence
CDS
ATGCAACCTTTATCAATGCATCCTAAGTCAGACATTCAGATTGATCTGACCCAGGTTAGAATTGATATTCTTCAAGGTATCCGTGAATGTAACAGTCGTGGTCTTGTACAAACTACGAAGTGGCTTGCAGAATTGAATTATGCTTTAAGAGACCACAAGATAACTTCCGAGGAACCTTCAGAATCTCTGAACGAGGACATAACGCCGGAGGAAAAGGATGCCTACACATTAGCAAAATGCTACTTCGACTGTCAAGAATACGATAGAGCTGCCCATTTTTTAGAGAACTGCACGAGTTCGAAATGCGTTTTTCTTCACAAATATTCTTTGTATATGTCTAGTGAGAAAAAGCGTTTAGATAATGCCACAGACCTGGGTACAGAAAATAGTGAATCAAATCAAGTTCTTTTGGACTTATTGTCTTTCTTTAAAACCAACTCCAATAACCTCGATGGTTACTTGTTGTATCTAGAAGGTGTTGTACTTAAAAAACTAGATCTAAGAAGTCAAGCTGTTACAGTACTGCAAGCTTCTGTTCAAGCAGCACCTACATTATGGGCAGCCTGGATAGAGTTGGCAGGCTTGGCCAATGAATATGAGGCACTAGATTCATTACAACTACCGAAACATTGGATGATGTACTTTTTTGCAGCTCATGCATTTGTGGAACTGAAGCTATCAGAACAAGCTTTGGAAGCTTACATGGTGTTGGCTGCAGCAGGATTTGAGAAGAGTACATATATAACAGCACAAATGGCTATAGCGCATCATGACAGAAGAGATGTGGATTCTTCACTTGCTTTGTTCAGGTTATTATACGAGAATGATCCATATCGACTAGACAACTGGGATGTTTACTCTCATCTGTTATATTTGAAGGAAAAACGAATGGAACTTGCAAATTTGGCACAAAGGGCAGTCTCCATTGACAAATACAGAGTAGAAACGTGTTGTGTTATAGGTAATTATTACAGCTTAAGATGTGAGCACCAAAAAGCTGTGATATACTTCCAAAGAGCTTTATCTCTAGACCCTCAGTACTTGTCTGCGTGGATCCTTATGGGGCATGAATTCATTGAACTTCAAAACTCCAATGCTGCAATACAATGTTATAGGCAAGCCATTGATGTAAACCGAAATGATTATAGAGCATGGAACGGTCTAGGACAAGCATATGAAATACTAGGGTTGAATAGTTACTGTATTTACTATTACTCTAGAGCCGCTCAACTGAAACCTGATGATTCAAGGATGTTGGTGTCTCTCGGTGAAGCATATGAGAAAATGGAAAAAATACCTAATGCATTGAAATGTTATTACAAAGCGCACAGTACAGGCGATATTGAAGGCATGGCTTTATTCAAATTAGCCAAGTTGTATGAAAGGTCGAACATGCCAAACAGTGCTGCGGCTGCGTACACAGCGGCGTGCCAGGATCCCGCGAACTCCGGCAGCAAGGAACTGCCGGCGGCGCAACGGTACCTAGCGCAGTATTACCTTAGGTTCTCACTGCTTGATCACGCGGCACATTACGCCTATAAATGTTTGGAGCATGAGAGTACTAAAGAAGCAGGAAAGAACATATTAAAAATGGTTTCCGAAAAACGGTTAGCAGCGTCTTCGTCCCAGTCGTCAGATTTACCTGAAGACTTACCAGAGGCAATCAAGAATGTGTCAACGGTGTCCATAACACAGGACATTCCCAAAACACCGCTACCTAGTCCCAATGTTACTCCAGAGAACTTCTCGACCCCGGTTTTCACGCGAAAAAACAAGCTTTGA
Protein
MQPLSMHPKSDIQIDLTQVRIDILQGIRECNSRGLVQTTKWLAELNYALRDHKITSEEPSESLNEDITPEEKDAYTLAKCYFDCQEYDRAAHFLENCTSSKCVFLHKYSLYMSSEKKRLDNATDLGTENSESNQVLLDLLSFFKTNSNNLDGYLLYLEGVVLKKLDLRSQAVTVLQASVQAAPTLWAAWIELAGLANEYEALDSLQLPKHWMMYFFAAHAFVELKLSEQALEAYMVLAAAGFEKSTYITAQMAIAHHDRRDVDSSLALFRLLYENDPYRLDNWDVYSHLLYLKEKRMELANLAQRAVSIDKYRVETCCVIGNYYSLRCEHQKAVIYFQRALSLDPQYLSAWILMGHEFIELQNSNAAIQCYRQAIDVNRNDYRAWNGLGQAYEILGLNSYCIYYYSRAAQLKPDDSRMLVSLGEAYEKMEKIPNALKCYYKAHSTGDIEGMALFKLAKLYERSNMPNSAAAAYTAACQDPANSGSKELPAAQRYLAQYYLRFSLLDHAAHYAYKCLEHESTKEAGKNILKMVSEKRLAASSSQSSDLPEDLPEAIKNVSTVSITQDIPKTPLPSPNVTPENFSTPVFTRKNKL
Summary
Uniprot
H9J0T7
A0A2A4JB81
A0A2H1VJB3
A0A212FFD5
A0A194PU28
A0A194R8Q7
+ More
A0A0L7LAE8 S4PM00 A0A2J7QCT7 A0A1W4WVK0 A0A1Y1LWT3 A0A067RJF3 E2C5D1 A0A3L8E1C3 A0A232F7T2 E2A0K5 A0A158NDN8 K7IP99 A0A0C9R501 A0A195CL96 A0A0J7KCT9 A0A195BQT6 A0A151XEJ9 A0A195F3U6 A0A2R5LLS6 A0A2A3E6H7 A0A088A2M6 F4WZ39 A0A0L7RBB5 A0A195D9X9 A0A293M7K3 A0A182GYE2 D6WM66 Q17FZ7 A0A1I8P8G1 A0A0M9A157 B0WWM2 A0A154PGG4 W5JD92 A0A131XZH7 A0A1B6J7D1 A0A1B6F0D6 A0A0A9XSR5 B4MZA8 A0A0V0GAC5 T1JH63 A0A1I8MBF9 A0A224XMW7 A0A2G8L161 A0A069DVK1 B7QK51 A0A1Q3F4J0 A0A0L0CAB3 A0A1W4V1F1 A0A182J1T8 A0A182F2I1 A0A1S3IXC3 B3MMU3 A0A084WC76 B4Q3D6 B4LSY4 B4KEP8 B4IFC4 Q9I7L8 B8A417 A0A0M4EP48 A0A0A1WEV5 B3NM12 A0A0P4WCT9 B4GSV7 A0A1A9XZI4 A0A1B0B909 Q29KS5 B4JCD1 A0A034WGC2 A0A1A9UMH9 A0A1B0FIR9 A0A182MFV3 A0A2P6KFY2 W8B7K7 A0A182X797 A0A182TZI3 A0A182KTM8 Q7Q1A6 A0A182PJI1 A0A3B0KDR3 B4PBL0 A0A210PPG0 C3ZLP9 A0A182HRK3 A0A182K3S4 A0A087U6G2 A0A0K8WDJ4 A0A182RAS1 A0A3B4GFC7 A0A3P9QJ58 A0A182YDN0 A0A182NUJ0 H3BCT6
A0A0L7LAE8 S4PM00 A0A2J7QCT7 A0A1W4WVK0 A0A1Y1LWT3 A0A067RJF3 E2C5D1 A0A3L8E1C3 A0A232F7T2 E2A0K5 A0A158NDN8 K7IP99 A0A0C9R501 A0A195CL96 A0A0J7KCT9 A0A195BQT6 A0A151XEJ9 A0A195F3U6 A0A2R5LLS6 A0A2A3E6H7 A0A088A2M6 F4WZ39 A0A0L7RBB5 A0A195D9X9 A0A293M7K3 A0A182GYE2 D6WM66 Q17FZ7 A0A1I8P8G1 A0A0M9A157 B0WWM2 A0A154PGG4 W5JD92 A0A131XZH7 A0A1B6J7D1 A0A1B6F0D6 A0A0A9XSR5 B4MZA8 A0A0V0GAC5 T1JH63 A0A1I8MBF9 A0A224XMW7 A0A2G8L161 A0A069DVK1 B7QK51 A0A1Q3F4J0 A0A0L0CAB3 A0A1W4V1F1 A0A182J1T8 A0A182F2I1 A0A1S3IXC3 B3MMU3 A0A084WC76 B4Q3D6 B4LSY4 B4KEP8 B4IFC4 Q9I7L8 B8A417 A0A0M4EP48 A0A0A1WEV5 B3NM12 A0A0P4WCT9 B4GSV7 A0A1A9XZI4 A0A1B0B909 Q29KS5 B4JCD1 A0A034WGC2 A0A1A9UMH9 A0A1B0FIR9 A0A182MFV3 A0A2P6KFY2 W8B7K7 A0A182X797 A0A182TZI3 A0A182KTM8 Q7Q1A6 A0A182PJI1 A0A3B0KDR3 B4PBL0 A0A210PPG0 C3ZLP9 A0A182HRK3 A0A182K3S4 A0A087U6G2 A0A0K8WDJ4 A0A182RAS1 A0A3B4GFC7 A0A3P9QJ58 A0A182YDN0 A0A182NUJ0 H3BCT6
Pubmed
19121390
22118469
26354079
26227816
23622113
28004739
+ More
24845553 20798317 30249741 28648823 21347285 20075255 21719571 26483478 18362917 19820115 17510324 20920257 23761445 25401762 26823975 17994087 25315136 29023486 26334808 26108605 24438588 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 15632085 25348373 24495485 20966253 12364791 17550304 28812685 18563158 25244985 9215903
24845553 20798317 30249741 28648823 21347285 20075255 21719571 26483478 18362917 19820115 17510324 20920257 23761445 25401762 26823975 17994087 25315136 29023486 26334808 26108605 24438588 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 15632085 25348373 24495485 20966253 12364791 17550304 28812685 18563158 25244985 9215903
EMBL
BABH01010322
NWSH01002294
PCG68653.1
ODYU01002883
SOQ40920.1
AGBW02008830
+ More
OWR52433.1 KQ459598 KPI94640.1 KQ460779 KPJ12256.1 JTDY01001995 KOB72374.1 GAIX01003790 JAA88770.1 NEVH01015876 PNF26398.1 GEZM01045001 JAV77989.1 KK852595 KDR20623.1 GL452770 EFN76864.1 QOIP01000002 RLU25828.1 NNAY01000743 OXU26725.1 GL435626 EFN72908.1 ADTU01012729 GBYB01011389 JAG81156.1 KQ977600 KYN01496.1 LBMM01009562 KMQ88026.1 KQ976424 KYM88324.1 KQ982254 KYQ58793.1 KQ981820 KYN35133.1 GGLE01006269 MBY10395.1 KZ288367 PBC26826.1 GL888465 EGI60542.1 KQ414617 KOC68110.1 KQ981082 KYN09725.1 GFWV01011352 MAA36081.1 JXUM01097358 KQ564337 KXJ72407.1 KQ971343 EFA04231.1 CH477267 EAT45473.1 KQ435793 KOX74256.1 DS232149 EDS36114.1 KQ434899 KZC10933.1 ADMH02001853 ETN60860.1 GEFM01004138 JAP71658.1 GECU01012623 JAS95083.1 GECZ01026119 JAS43650.1 GBHO01020595 GBRD01011484 GDHC01016702 JAG23009.1 JAG54340.1 JAQ01927.1 CH963913 EDW77381.2 GECL01001115 JAP05009.1 JH432222 GFTR01007087 JAW09339.1 MRZV01000267 PIK54004.1 GBGD01001058 JAC87831.1 ABJB010360414 DS957045 EEC19223.1 GFDL01012571 JAV22474.1 JRES01000682 KNC29175.1 CH902620 EDV30968.1 ATLV01022579 KE525333 KFB47820.1 CM000361 CM002910 EDX05617.1 KMY91166.1 CH940649 EDW64895.1 KRF81928.1 KRF81929.1 CH933807 EDW12948.1 CH480833 EDW46346.1 AE014134 BT010078 AAG22447.2 AAQ22547.1 AGB93164.1 BT056309 ACL68756.1 CP012523 ALC38562.1 GBXI01016870 JAC97421.1 CH954179 EDV54548.1 GDRN01099305 JAI58793.1 CH479189 EDW25466.1 JXJN01010202 CH379061 EAL33100.1 CH916368 EDW04164.1 GAKP01006134 JAC52818.1 CCAG010016358 AXCM01000820 MWRG01013076 PRD25217.1 GAMC01011978 JAB94577.1 AAAB01008980 EAA14469.3 OUUW01000006 SPP81798.1 CM000158 EDW90524.1 NEDP02005569 OWF38363.1 GG666642 EEN46522.1 APCN01001715 KK118433 KFM72951.1 GDHF01003137 JAI49177.1 AFYH01039562
OWR52433.1 KQ459598 KPI94640.1 KQ460779 KPJ12256.1 JTDY01001995 KOB72374.1 GAIX01003790 JAA88770.1 NEVH01015876 PNF26398.1 GEZM01045001 JAV77989.1 KK852595 KDR20623.1 GL452770 EFN76864.1 QOIP01000002 RLU25828.1 NNAY01000743 OXU26725.1 GL435626 EFN72908.1 ADTU01012729 GBYB01011389 JAG81156.1 KQ977600 KYN01496.1 LBMM01009562 KMQ88026.1 KQ976424 KYM88324.1 KQ982254 KYQ58793.1 KQ981820 KYN35133.1 GGLE01006269 MBY10395.1 KZ288367 PBC26826.1 GL888465 EGI60542.1 KQ414617 KOC68110.1 KQ981082 KYN09725.1 GFWV01011352 MAA36081.1 JXUM01097358 KQ564337 KXJ72407.1 KQ971343 EFA04231.1 CH477267 EAT45473.1 KQ435793 KOX74256.1 DS232149 EDS36114.1 KQ434899 KZC10933.1 ADMH02001853 ETN60860.1 GEFM01004138 JAP71658.1 GECU01012623 JAS95083.1 GECZ01026119 JAS43650.1 GBHO01020595 GBRD01011484 GDHC01016702 JAG23009.1 JAG54340.1 JAQ01927.1 CH963913 EDW77381.2 GECL01001115 JAP05009.1 JH432222 GFTR01007087 JAW09339.1 MRZV01000267 PIK54004.1 GBGD01001058 JAC87831.1 ABJB010360414 DS957045 EEC19223.1 GFDL01012571 JAV22474.1 JRES01000682 KNC29175.1 CH902620 EDV30968.1 ATLV01022579 KE525333 KFB47820.1 CM000361 CM002910 EDX05617.1 KMY91166.1 CH940649 EDW64895.1 KRF81928.1 KRF81929.1 CH933807 EDW12948.1 CH480833 EDW46346.1 AE014134 BT010078 AAG22447.2 AAQ22547.1 AGB93164.1 BT056309 ACL68756.1 CP012523 ALC38562.1 GBXI01016870 JAC97421.1 CH954179 EDV54548.1 GDRN01099305 JAI58793.1 CH479189 EDW25466.1 JXJN01010202 CH379061 EAL33100.1 CH916368 EDW04164.1 GAKP01006134 JAC52818.1 CCAG010016358 AXCM01000820 MWRG01013076 PRD25217.1 GAMC01011978 JAB94577.1 AAAB01008980 EAA14469.3 OUUW01000006 SPP81798.1 CM000158 EDW90524.1 NEDP02005569 OWF38363.1 GG666642 EEN46522.1 APCN01001715 KK118433 KFM72951.1 GDHF01003137 JAI49177.1 AFYH01039562
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000235965 UP000192223 UP000027135 UP000008237 UP000279307 UP000215335 UP000000311 UP000005205 UP000002358 UP000078542 UP000036403 UP000078540 UP000075809 UP000078541 UP000242457 UP000005203 UP000007755 UP000053825 UP000078492 UP000069940 UP000249989 UP000007266 UP000008820 UP000095300 UP000053105 UP000002320 UP000076502 UP000000673 UP000007798 UP000095301 UP000230750 UP000001555 UP000037069 UP000192221 UP000075880 UP000069272 UP000085678 UP000007801 UP000030765 UP000000304 UP000008792 UP000009192 UP000001292 UP000000803 UP000092553 UP000008711 UP000008744 UP000092443 UP000092460 UP000001819 UP000001070 UP000078200 UP000092444 UP000075883 UP000076407 UP000075902 UP000075882 UP000007062 UP000075885 UP000268350 UP000002282 UP000242188 UP000001554 UP000075840 UP000075881 UP000054359 UP000075900 UP000261460 UP000242638 UP000076408 UP000075884 UP000008672
UP000235965 UP000192223 UP000027135 UP000008237 UP000279307 UP000215335 UP000000311 UP000005205 UP000002358 UP000078542 UP000036403 UP000078540 UP000075809 UP000078541 UP000242457 UP000005203 UP000007755 UP000053825 UP000078492 UP000069940 UP000249989 UP000007266 UP000008820 UP000095300 UP000053105 UP000002320 UP000076502 UP000000673 UP000007798 UP000095301 UP000230750 UP000001555 UP000037069 UP000192221 UP000075880 UP000069272 UP000085678 UP000007801 UP000030765 UP000000304 UP000008792 UP000009192 UP000001292 UP000000803 UP000092553 UP000008711 UP000008744 UP000092443 UP000092460 UP000001819 UP000001070 UP000078200 UP000092444 UP000075883 UP000076407 UP000075902 UP000075882 UP000007062 UP000075885 UP000268350 UP000002282 UP000242188 UP000001554 UP000075840 UP000075881 UP000054359 UP000075900 UP000261460 UP000242638 UP000076408 UP000075884 UP000008672
Interpro
Gene 3D
ProteinModelPortal
H9J0T7
A0A2A4JB81
A0A2H1VJB3
A0A212FFD5
A0A194PU28
A0A194R8Q7
+ More
A0A0L7LAE8 S4PM00 A0A2J7QCT7 A0A1W4WVK0 A0A1Y1LWT3 A0A067RJF3 E2C5D1 A0A3L8E1C3 A0A232F7T2 E2A0K5 A0A158NDN8 K7IP99 A0A0C9R501 A0A195CL96 A0A0J7KCT9 A0A195BQT6 A0A151XEJ9 A0A195F3U6 A0A2R5LLS6 A0A2A3E6H7 A0A088A2M6 F4WZ39 A0A0L7RBB5 A0A195D9X9 A0A293M7K3 A0A182GYE2 D6WM66 Q17FZ7 A0A1I8P8G1 A0A0M9A157 B0WWM2 A0A154PGG4 W5JD92 A0A131XZH7 A0A1B6J7D1 A0A1B6F0D6 A0A0A9XSR5 B4MZA8 A0A0V0GAC5 T1JH63 A0A1I8MBF9 A0A224XMW7 A0A2G8L161 A0A069DVK1 B7QK51 A0A1Q3F4J0 A0A0L0CAB3 A0A1W4V1F1 A0A182J1T8 A0A182F2I1 A0A1S3IXC3 B3MMU3 A0A084WC76 B4Q3D6 B4LSY4 B4KEP8 B4IFC4 Q9I7L8 B8A417 A0A0M4EP48 A0A0A1WEV5 B3NM12 A0A0P4WCT9 B4GSV7 A0A1A9XZI4 A0A1B0B909 Q29KS5 B4JCD1 A0A034WGC2 A0A1A9UMH9 A0A1B0FIR9 A0A182MFV3 A0A2P6KFY2 W8B7K7 A0A182X797 A0A182TZI3 A0A182KTM8 Q7Q1A6 A0A182PJI1 A0A3B0KDR3 B4PBL0 A0A210PPG0 C3ZLP9 A0A182HRK3 A0A182K3S4 A0A087U6G2 A0A0K8WDJ4 A0A182RAS1 A0A3B4GFC7 A0A3P9QJ58 A0A182YDN0 A0A182NUJ0 H3BCT6
A0A0L7LAE8 S4PM00 A0A2J7QCT7 A0A1W4WVK0 A0A1Y1LWT3 A0A067RJF3 E2C5D1 A0A3L8E1C3 A0A232F7T2 E2A0K5 A0A158NDN8 K7IP99 A0A0C9R501 A0A195CL96 A0A0J7KCT9 A0A195BQT6 A0A151XEJ9 A0A195F3U6 A0A2R5LLS6 A0A2A3E6H7 A0A088A2M6 F4WZ39 A0A0L7RBB5 A0A195D9X9 A0A293M7K3 A0A182GYE2 D6WM66 Q17FZ7 A0A1I8P8G1 A0A0M9A157 B0WWM2 A0A154PGG4 W5JD92 A0A131XZH7 A0A1B6J7D1 A0A1B6F0D6 A0A0A9XSR5 B4MZA8 A0A0V0GAC5 T1JH63 A0A1I8MBF9 A0A224XMW7 A0A2G8L161 A0A069DVK1 B7QK51 A0A1Q3F4J0 A0A0L0CAB3 A0A1W4V1F1 A0A182J1T8 A0A182F2I1 A0A1S3IXC3 B3MMU3 A0A084WC76 B4Q3D6 B4LSY4 B4KEP8 B4IFC4 Q9I7L8 B8A417 A0A0M4EP48 A0A0A1WEV5 B3NM12 A0A0P4WCT9 B4GSV7 A0A1A9XZI4 A0A1B0B909 Q29KS5 B4JCD1 A0A034WGC2 A0A1A9UMH9 A0A1B0FIR9 A0A182MFV3 A0A2P6KFY2 W8B7K7 A0A182X797 A0A182TZI3 A0A182KTM8 Q7Q1A6 A0A182PJI1 A0A3B0KDR3 B4PBL0 A0A210PPG0 C3ZLP9 A0A182HRK3 A0A182K3S4 A0A087U6G2 A0A0K8WDJ4 A0A182RAS1 A0A3B4GFC7 A0A3P9QJ58 A0A182YDN0 A0A182NUJ0 H3BCT6
PDB
5LCW
E-value=1.27739e-134,
Score=1231
Ontologies
GO
Topology
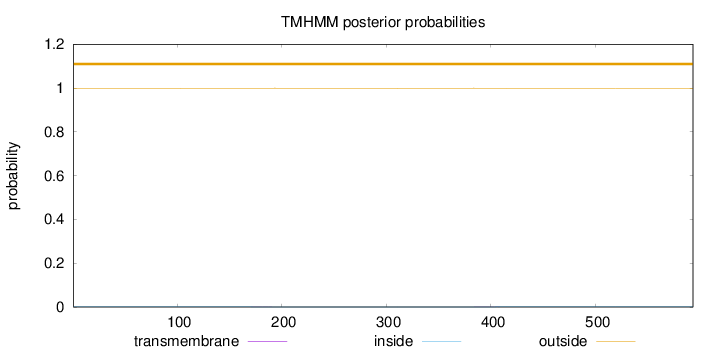
Length:
593
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01719
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00076
outside
1 - 593
Population Genetic Test Statistics
Pi
270.430983
Theta
197.007262
Tajima's D
1.351365
CLR
0
CSRT
0.746712664366782
Interpretation
Uncertain
