Gene
KWMTBOMO02200
Pre Gene Modal
BGIBMGA003033
Annotation
PREDICTED:_asparagine_synthetase_domain-containing_protein_CG17486_isoform_X1_[Bombyx_mori]
Full name
Asparagine synthetase domain-containing protein CG17486
Location in the cell
Nuclear Reliability : 3.697
Sequence
CDS
ATGTGTGGTATATTTTTGGAGTTAAGCTGCGAAAAATGTAGCATAGTGAACGATATTGATGTCCTTCGAAGACTCAAAAACAGAGGACCCGATCACTTACAGAGGAAGGATTTTATTTCTGAAATTGGACACCACATAGTTTTCGTAGCCTCAGTTCTATGGATGCAAGGTCCGCAAATGACATTACAACCTCTCGAAAATGAATGTGGCATCCTTTTGTATAATGGTGATATATTTGACGAATCTTGGGATAGCAGAACCAGCGACACGCAAATAATCATGGAGAAACTTAGTTATGATTGTGACAGTCAAATCGATAAAGTGATCAATGTATTAAAGTCACTAAAGGGGCCATTCAGCTTAATATATTACTGTAAAGTGACAAATAAATTATATTTCACTAGGGATCGATTTGGAAGAAACTCTTTACTCATACATAAAAATTCAAATTCTTTAGTATTATCCAGTGTCTTAGGTACACAGTATGAGAATTATGAAGTTCCCGTATCACACATCTCTGTCTTGGACTTGAATACTCATAAGATATTGTTGTATACATGGAAAGCCACAGATATCGTTCAGCAATGTAATCTCAGTGATTGGTTGAAGACATTACAATTAAAACAAAGTTTATCTGATGAAGATTTTACCGTTGAAGCCAATATACCATGGGATATAGATGGTCTTGAGGATACAGTGTCATTCATTGAAAGTGTTGCTAAGAAAACAAAATGTAATGTAACTATTATGAAGACGCTTAGTCAACACCCTGAAATACAAACAACAATTAATAAAATAATTTACTTACTAAATAAAAGTATTCAGAAAAGGTTAAAACGCCAGCCAAATTTGTGTAGGAATTGCTTGAAAATTACAGAAATAAACTGTAGTCATAGTGCGGTTGGAGTATTATTTTCGGGTGGCTTGGATTGTACAATACTTGCATTGCTAGCTGATAAGTATGTACCAAAAAACCAAAGCATAGATTTGCTTAATGTAGCATTTAATAAAGAGAACCCAACATATGATGTACCTGACAGGATAACCGGTAGAAAATCTTTAGAAGAGCTAAGAAGATTATGTCCATTACGGAAATGGACTTTCAAAGAAATTAATGTACCAAAAGAAGAGTTGGAAGAATTTCAGATTAATGTAATAGCTGACTTGATTTATCCAAGACAAACTATTCTGGATGAAAGTCTTGGCTCAGCATTATGGTTTGCCGCTCGAGGCCAAGACCATCACTCCGTGTCTAATTGTAGGGTTTTATTTAGTGGTACTGGTGCAGATGAATTATTTGGTGGATACACAAGACACAGGGCTGTTTTTAAACGGTTAGGATGGGCTGGTCTATGTAAAGAATTACAATTAGATTGGGAAAGGATCCCATACAGAAATTTAGCTCGGGATAACAGAGTTATTTGTGATCATGGTCGACAACCCAGAATGCCCTATCTAGATGAAGAATTTGCTGAATTTGTTATCCATTTAAAACCTTGGTTAAAATGTTTTCCAAGTAAACATTTGGGCCATGGCTTAGGCGACAAATTAATGCTGAGACTCGTAGCTTATAATATTGGTTTGATGGAAGTCTCTACGTTTCCCAAAAGAGCTCTACAATTTGGATCTCGAGTAGCAAACAAGAATGAAAAAGGTTGTGATAAGTCATTTTTTTTCAGGAATCAAATAAAAAATAAAATAGTATCATGA
Protein
MCGIFLELSCEKCSIVNDIDVLRRLKNRGPDHLQRKDFISEIGHHIVFVASVLWMQGPQMTLQPLENECGILLYNGDIFDESWDSRTSDTQIIMEKLSYDCDSQIDKVINVLKSLKGPFSLIYYCKVTNKLYFTRDRFGRNSLLIHKNSNSLVLSSVLGTQYENYEVPVSHISVLDLNTHKILLYTWKATDIVQQCNLSDWLKTLQLKQSLSDEDFTVEANIPWDIDGLEDTVSFIESVAKKTKCNVTIMKTLSQHPEIQTTINKIIYLLNKSIQKRLKRQPNLCRNCLKITEINCSHSAVGVLFSGGLDCTILALLADKYVPKNQSIDLLNVAFNKENPTYDVPDRITGRKSLEELRRLCPLRKWTFKEINVPKEELEEFQINVIADLIYPRQTILDESLGSALWFAARGQDHHSVSNCRVLFSGTGADELFGGYTRHRAVFKRLGWAGLCKELQLDWERIPYRNLARDNRVICDHGRQPRMPYLDEEFAEFVIHLKPWLKCFPSKHLGHGLGDKLMLRLVAYNIGLMEVSTFPKRALQFGSRVANKNEKGCDKSFFFRNQIKNKIVS
Summary
Keywords
Amino-acid biosynthesis
Asparagine biosynthesis
Complete proteome
Glutamine amidotransferase
Reference proteome
Feature
chain Asparagine synthetase domain-containing protein CG17486
Uniprot
A0A2A4JAE2
A0A2H1VJF1
A0A194PMI0
A0A212FFB5
A0A0L7LA85
A0A194R3I5
+ More
J3JT65 A0A026W5F7 E2BLK8 A0A0J7KU69 A0A2A3E3R0 A0A1W4X6Y9 A0A182YFA2 D6WQE0 E2AIU4 A0A0C9PP67 A0A154PT22 A0A151IMR4 A0A195ELZ8 A0A1Y1KR92 U4UHE7 A0A195ESP0 A0A151X8I3 A0A195ATB5 A0A158NPQ3 A0A1S4FDJ0 Q176B2 F4W7I4 A0A182GHT7 E9J7T1 A0A1B0DHM8 A0A0L7R394 K7IWQ8 E0VSE8 A0A182NDD6 A0A0M8ZSW0 A0A182PUH0 N6UBZ2 A0A1S6KZL6 A0A088AIS4 A0A182RIU9 A0NDZ3 A0A182HYG7 A0A182LPW2 A0A1W4V863 A0A084WAJ3 A0A224XLE6 A0A023F173 A0A182JQQ4 B0WP11 A0A0K8T8K4 A0A1B6D251 A0A0A9ZE66 A0A146MF32 A0A2H8TE83 A0A182WUX2 A0A0J9R5S5 A0A310SFG4 A0A0R1E7X4 A0A0Q5WLM0 W8C3P6 A0A0L0BL09 A0A162SX85 Q5LJP9 A0A0P5XL66 A0A0P6GT01 A0A0P6J619 A0A0P5LBN2 J9JRN3 A0A0P6B8F9 B5DSC1 A0A0P5RWQ8 A0A0N8B8X1 A0A1Y1KRI4 A0A0P5YMS5 A0A293LFK4 A0A2L2YMF4 A0A1B0AZC5 A0A336LXQ2 A0A1I8Q8J1 A0A1I8Q8F8 A0A1Z5L1V7 B4M753 A0A0P6D081 A0A0P5H5X6 E9H1C3 B4JX80 W5JNQ9 A0A087T016 T1HBW4 B4NN67 B4L5Z8 A0A0K2UQD1 T1JEG1 A0A232EZ62 A0A0M4EI64 A0A0A1X8Z5 A0A0P8XKN7
J3JT65 A0A026W5F7 E2BLK8 A0A0J7KU69 A0A2A3E3R0 A0A1W4X6Y9 A0A182YFA2 D6WQE0 E2AIU4 A0A0C9PP67 A0A154PT22 A0A151IMR4 A0A195ELZ8 A0A1Y1KR92 U4UHE7 A0A195ESP0 A0A151X8I3 A0A195ATB5 A0A158NPQ3 A0A1S4FDJ0 Q176B2 F4W7I4 A0A182GHT7 E9J7T1 A0A1B0DHM8 A0A0L7R394 K7IWQ8 E0VSE8 A0A182NDD6 A0A0M8ZSW0 A0A182PUH0 N6UBZ2 A0A1S6KZL6 A0A088AIS4 A0A182RIU9 A0NDZ3 A0A182HYG7 A0A182LPW2 A0A1W4V863 A0A084WAJ3 A0A224XLE6 A0A023F173 A0A182JQQ4 B0WP11 A0A0K8T8K4 A0A1B6D251 A0A0A9ZE66 A0A146MF32 A0A2H8TE83 A0A182WUX2 A0A0J9R5S5 A0A310SFG4 A0A0R1E7X4 A0A0Q5WLM0 W8C3P6 A0A0L0BL09 A0A162SX85 Q5LJP9 A0A0P5XL66 A0A0P6GT01 A0A0P6J619 A0A0P5LBN2 J9JRN3 A0A0P6B8F9 B5DSC1 A0A0P5RWQ8 A0A0N8B8X1 A0A1Y1KRI4 A0A0P5YMS5 A0A293LFK4 A0A2L2YMF4 A0A1B0AZC5 A0A336LXQ2 A0A1I8Q8J1 A0A1I8Q8F8 A0A1Z5L1V7 B4M753 A0A0P6D081 A0A0P5H5X6 E9H1C3 B4JX80 W5JNQ9 A0A087T016 T1HBW4 B4NN67 B4L5Z8 A0A0K2UQD1 T1JEG1 A0A232EZ62 A0A0M4EI64 A0A0A1X8Z5 A0A0P8XKN7
Pubmed
26354079
22118469
26227816
22516182
24508170
30249741
+ More
20798317 25244985 18362917 19820115 28004739 23537049 21347285 17510324 21719571 26483478 21282665 20075255 20566863 24330026 12364791 20966253 24438588 25474469 25401762 26823975 22936249 17994087 17550304 24495485 26108605 10731132 12537572 12537569 15632085 26561354 28528879 21292972 20920257 23761445 28648823 25830018
20798317 25244985 18362917 19820115 28004739 23537049 21347285 17510324 21719571 26483478 21282665 20075255 20566863 24330026 12364791 20966253 24438588 25474469 25401762 26823975 22936249 17994087 17550304 24495485 26108605 10731132 12537572 12537569 15632085 26561354 28528879 21292972 20920257 23761445 28648823 25830018
EMBL
NWSH01002294
PCG68648.1
ODYU01002883
SOQ40921.1
KQ459598
KPI94641.1
+ More
AGBW02008830 OWR52435.1 JTDY01001995 KOB72377.1 KQ460779 KPJ12257.1 BT126421 AEE61385.1 KK107467 QOIP01000013 EZA50264.1 RLU15542.1 GL449036 EFN83342.1 LBMM01003167 KMQ93868.1 KZ288408 PBC26124.1 KQ971354 EFA07688.2 GL439905 EFN66623.1 GBYB01002978 JAG72745.1 KQ435207 KZC15065.1 KQ977026 KYN06245.1 KQ978730 KYN28947.1 GEZM01075843 JAV63923.1 KB632326 ERL92437.1 KQ981986 KYN31248.1 KQ982409 KYQ56686.1 KQ976745 KYM75436.1 ADTU01022707 ADTU01022708 CH477391 EAT41946.1 GL887844 EGI69857.1 JXUM01064744 JXUM01064745 JXUM01064746 JXUM01064747 KQ562313 KXJ76164.1 GL768630 EFZ11121.1 AJVK01061335 KQ414663 KOC65337.1 AAZX01000616 DS235750 EEB16304.1 KQ435911 KOX68943.1 APGK01035026 APGK01035027 KB740923 ENN78151.1 KC223371 AQT27180.1 AAAB01008933 EAU76749.2 APCN01005907 ATLV01022200 KE525330 KFB47237.1 GFTR01007176 JAW09250.1 GBBI01003941 JAC14771.1 DS232018 EDS32038.1 GBRD01004346 JAG61475.1 GEDC01017572 JAS19726.1 GBHO01001408 JAG42196.1 GDHC01001017 JAQ17612.1 GFXV01000596 MBW12401.1 CM002911 KMY91557.1 KQ769179 OAD52969.1 CH891612 KRK05141.1 CH954177 KQS71070.1 GAMC01010026 JAB96529.1 JRES01001702 KNC20790.1 LRGB01000024 KZS21706.1 AE013599 AY069456 GDIP01070493 JAM33222.1 GDIQ01030286 JAN64451.1 GDIQ01022768 JAN71969.1 GDIQ01171853 JAK79872.1 ABLF02011554 GDIP01024958 JAM78757.1 CH475486 EDY70881.1 GDIQ01095245 JAL56481.1 GDIQ01201291 JAK50434.1 GEZM01075842 JAV63924.1 GDIP01055683 JAM48032.1 GFWV01002783 MAA27513.1 IAAA01034848 LAA08555.1 JXJN01006237 UFQS01000038 UFQT01000038 SSW98052.1 SSX18438.1 GFJQ02005695 JAW01275.1 CH940653 EDW62620.1 GDIQ01087130 JAN07607.1 GDIQ01237656 JAK14069.1 GL732583 EFX74444.1 CH916376 EDV95356.1 ADMH02000540 ETN66017.1 KK112757 KFM58455.1 ACPB03010887 CH964282 EDW85806.2 CH933812 EDW05794.2 HACA01022580 CDW39941.1 JH432117 NNAY01001537 OXU23661.1 CP012528 ALC48504.1 GBXI01006690 JAD07602.1 CH902628 KPU75341.1
AGBW02008830 OWR52435.1 JTDY01001995 KOB72377.1 KQ460779 KPJ12257.1 BT126421 AEE61385.1 KK107467 QOIP01000013 EZA50264.1 RLU15542.1 GL449036 EFN83342.1 LBMM01003167 KMQ93868.1 KZ288408 PBC26124.1 KQ971354 EFA07688.2 GL439905 EFN66623.1 GBYB01002978 JAG72745.1 KQ435207 KZC15065.1 KQ977026 KYN06245.1 KQ978730 KYN28947.1 GEZM01075843 JAV63923.1 KB632326 ERL92437.1 KQ981986 KYN31248.1 KQ982409 KYQ56686.1 KQ976745 KYM75436.1 ADTU01022707 ADTU01022708 CH477391 EAT41946.1 GL887844 EGI69857.1 JXUM01064744 JXUM01064745 JXUM01064746 JXUM01064747 KQ562313 KXJ76164.1 GL768630 EFZ11121.1 AJVK01061335 KQ414663 KOC65337.1 AAZX01000616 DS235750 EEB16304.1 KQ435911 KOX68943.1 APGK01035026 APGK01035027 KB740923 ENN78151.1 KC223371 AQT27180.1 AAAB01008933 EAU76749.2 APCN01005907 ATLV01022200 KE525330 KFB47237.1 GFTR01007176 JAW09250.1 GBBI01003941 JAC14771.1 DS232018 EDS32038.1 GBRD01004346 JAG61475.1 GEDC01017572 JAS19726.1 GBHO01001408 JAG42196.1 GDHC01001017 JAQ17612.1 GFXV01000596 MBW12401.1 CM002911 KMY91557.1 KQ769179 OAD52969.1 CH891612 KRK05141.1 CH954177 KQS71070.1 GAMC01010026 JAB96529.1 JRES01001702 KNC20790.1 LRGB01000024 KZS21706.1 AE013599 AY069456 GDIP01070493 JAM33222.1 GDIQ01030286 JAN64451.1 GDIQ01022768 JAN71969.1 GDIQ01171853 JAK79872.1 ABLF02011554 GDIP01024958 JAM78757.1 CH475486 EDY70881.1 GDIQ01095245 JAL56481.1 GDIQ01201291 JAK50434.1 GEZM01075842 JAV63924.1 GDIP01055683 JAM48032.1 GFWV01002783 MAA27513.1 IAAA01034848 LAA08555.1 JXJN01006237 UFQS01000038 UFQT01000038 SSW98052.1 SSX18438.1 GFJQ02005695 JAW01275.1 CH940653 EDW62620.1 GDIQ01087130 JAN07607.1 GDIQ01237656 JAK14069.1 GL732583 EFX74444.1 CH916376 EDV95356.1 ADMH02000540 ETN66017.1 KK112757 KFM58455.1 ACPB03010887 CH964282 EDW85806.2 CH933812 EDW05794.2 HACA01022580 CDW39941.1 JH432117 NNAY01001537 OXU23661.1 CP012528 ALC48504.1 GBXI01006690 JAD07602.1 CH902628 KPU75341.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000037510
UP000053240
UP000053097
+ More
UP000279307 UP000008237 UP000036403 UP000242457 UP000192223 UP000076408 UP000007266 UP000000311 UP000076502 UP000078542 UP000078492 UP000030742 UP000078541 UP000075809 UP000078540 UP000005205 UP000008820 UP000007755 UP000069940 UP000249989 UP000092462 UP000053825 UP000002358 UP000009046 UP000075884 UP000053105 UP000075885 UP000019118 UP000005203 UP000075900 UP000007062 UP000075840 UP000075882 UP000192221 UP000030765 UP000075881 UP000002320 UP000076407 UP000002282 UP000008711 UP000037069 UP000076858 UP000000803 UP000007819 UP000001819 UP000092460 UP000095300 UP000008792 UP000000305 UP000001070 UP000000673 UP000054359 UP000015103 UP000007798 UP000009192 UP000215335 UP000092553 UP000007801
UP000279307 UP000008237 UP000036403 UP000242457 UP000192223 UP000076408 UP000007266 UP000000311 UP000076502 UP000078542 UP000078492 UP000030742 UP000078541 UP000075809 UP000078540 UP000005205 UP000008820 UP000007755 UP000069940 UP000249989 UP000092462 UP000053825 UP000002358 UP000009046 UP000075884 UP000053105 UP000075885 UP000019118 UP000005203 UP000075900 UP000007062 UP000075840 UP000075882 UP000192221 UP000030765 UP000075881 UP000002320 UP000076407 UP000002282 UP000008711 UP000037069 UP000076858 UP000000803 UP000007819 UP000001819 UP000092460 UP000095300 UP000008792 UP000000305 UP000001070 UP000000673 UP000054359 UP000015103 UP000007798 UP000009192 UP000215335 UP000092553 UP000007801
PRIDE
Pfam
Interpro
IPR029055
Ntn_hydrolases_N
+ More
IPR017932 GATase_2_dom
IPR014729 Rossmann-like_a/b/a_fold
IPR001962 Asn_synthase
IPR037278 ARFGAP/RecO
IPR038508 ArfGAP_dom_sf
IPR001164 ArfGAP_dom
IPR006426 Asn_synth_AEB
IPR006751 TAFII55_prot_cons_reg
IPR001465 Malate_synthase
IPR006252 Malate_synthA
IPR011076 Malate_synth-like_sf
IPR017932 GATase_2_dom
IPR014729 Rossmann-like_a/b/a_fold
IPR001962 Asn_synthase
IPR037278 ARFGAP/RecO
IPR038508 ArfGAP_dom_sf
IPR001164 ArfGAP_dom
IPR006426 Asn_synth_AEB
IPR006751 TAFII55_prot_cons_reg
IPR001465 Malate_synthase
IPR006252 Malate_synthA
IPR011076 Malate_synth-like_sf
Gene 3D
ProteinModelPortal
A0A2A4JAE2
A0A2H1VJF1
A0A194PMI0
A0A212FFB5
A0A0L7LA85
A0A194R3I5
+ More
J3JT65 A0A026W5F7 E2BLK8 A0A0J7KU69 A0A2A3E3R0 A0A1W4X6Y9 A0A182YFA2 D6WQE0 E2AIU4 A0A0C9PP67 A0A154PT22 A0A151IMR4 A0A195ELZ8 A0A1Y1KR92 U4UHE7 A0A195ESP0 A0A151X8I3 A0A195ATB5 A0A158NPQ3 A0A1S4FDJ0 Q176B2 F4W7I4 A0A182GHT7 E9J7T1 A0A1B0DHM8 A0A0L7R394 K7IWQ8 E0VSE8 A0A182NDD6 A0A0M8ZSW0 A0A182PUH0 N6UBZ2 A0A1S6KZL6 A0A088AIS4 A0A182RIU9 A0NDZ3 A0A182HYG7 A0A182LPW2 A0A1W4V863 A0A084WAJ3 A0A224XLE6 A0A023F173 A0A182JQQ4 B0WP11 A0A0K8T8K4 A0A1B6D251 A0A0A9ZE66 A0A146MF32 A0A2H8TE83 A0A182WUX2 A0A0J9R5S5 A0A310SFG4 A0A0R1E7X4 A0A0Q5WLM0 W8C3P6 A0A0L0BL09 A0A162SX85 Q5LJP9 A0A0P5XL66 A0A0P6GT01 A0A0P6J619 A0A0P5LBN2 J9JRN3 A0A0P6B8F9 B5DSC1 A0A0P5RWQ8 A0A0N8B8X1 A0A1Y1KRI4 A0A0P5YMS5 A0A293LFK4 A0A2L2YMF4 A0A1B0AZC5 A0A336LXQ2 A0A1I8Q8J1 A0A1I8Q8F8 A0A1Z5L1V7 B4M753 A0A0P6D081 A0A0P5H5X6 E9H1C3 B4JX80 W5JNQ9 A0A087T016 T1HBW4 B4NN67 B4L5Z8 A0A0K2UQD1 T1JEG1 A0A232EZ62 A0A0M4EI64 A0A0A1X8Z5 A0A0P8XKN7
J3JT65 A0A026W5F7 E2BLK8 A0A0J7KU69 A0A2A3E3R0 A0A1W4X6Y9 A0A182YFA2 D6WQE0 E2AIU4 A0A0C9PP67 A0A154PT22 A0A151IMR4 A0A195ELZ8 A0A1Y1KR92 U4UHE7 A0A195ESP0 A0A151X8I3 A0A195ATB5 A0A158NPQ3 A0A1S4FDJ0 Q176B2 F4W7I4 A0A182GHT7 E9J7T1 A0A1B0DHM8 A0A0L7R394 K7IWQ8 E0VSE8 A0A182NDD6 A0A0M8ZSW0 A0A182PUH0 N6UBZ2 A0A1S6KZL6 A0A088AIS4 A0A182RIU9 A0NDZ3 A0A182HYG7 A0A182LPW2 A0A1W4V863 A0A084WAJ3 A0A224XLE6 A0A023F173 A0A182JQQ4 B0WP11 A0A0K8T8K4 A0A1B6D251 A0A0A9ZE66 A0A146MF32 A0A2H8TE83 A0A182WUX2 A0A0J9R5S5 A0A310SFG4 A0A0R1E7X4 A0A0Q5WLM0 W8C3P6 A0A0L0BL09 A0A162SX85 Q5LJP9 A0A0P5XL66 A0A0P6GT01 A0A0P6J619 A0A0P5LBN2 J9JRN3 A0A0P6B8F9 B5DSC1 A0A0P5RWQ8 A0A0N8B8X1 A0A1Y1KRI4 A0A0P5YMS5 A0A293LFK4 A0A2L2YMF4 A0A1B0AZC5 A0A336LXQ2 A0A1I8Q8J1 A0A1I8Q8F8 A0A1Z5L1V7 B4M753 A0A0P6D081 A0A0P5H5X6 E9H1C3 B4JX80 W5JNQ9 A0A087T016 T1HBW4 B4NN67 B4L5Z8 A0A0K2UQD1 T1JEG1 A0A232EZ62 A0A0M4EI64 A0A0A1X8Z5 A0A0P8XKN7
PDB
1CT9
E-value=3.72553e-06,
Score=123
Ontologies
GO
PANTHER
Topology
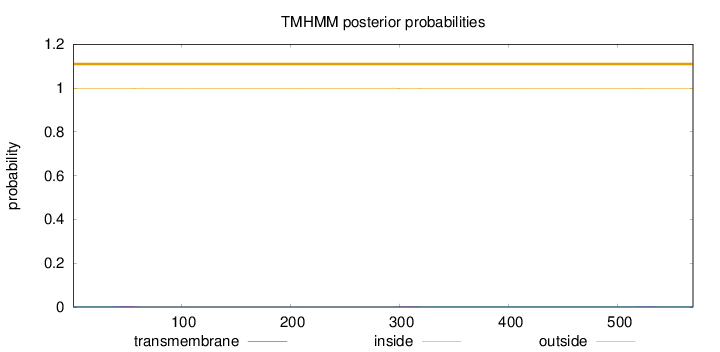
Length:
569
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10848
Exp number, first 60 AAs:
0.04882
Total prob of N-in:
0.00313
outside
1 - 569
Population Genetic Test Statistics
Pi
178.126846
Theta
177.923028
Tajima's D
-0.339609
CLR
0.932754
CSRT
0.284285785710714
Interpretation
Uncertain
