Pre Gene Modal
BGIBMGA003126
Annotation
PREDICTED:_nuclear_pore_complex_protein_Nup93-like_[Amyelois_transitella]
Full name
Nuclear pore protein
Location in the cell
Cytoplasmic Reliability : 2.234
Sequence
CDS
ATGGGAGATGAAGGATTTAGTAAACTACTTCAACAAGCAGAACAGTTAACGACTGAGATAGAAGGTAATGAAGAATTGCCGAGAGTTGAACGTTCTTTAGGGCAAGTTCTCGAGGCCTCCCAGGAACTTTACTCGAGAGTAATACAATCTGGCGCTAACGATATTCAAGCGCACTTATTGTTTGGCTCTAAAGGAATTGATCTAAACCAAATCTCACAAAAATTGGAGACACTTAGTTCAAAGCGTACTTTTGAGCCCCTCCAGCCAATAGCTGATTCAGATATTGAAAGTTTTTTGAAGAATGAAAAAGAAAATGCTATATTATCCCTTATTGATGAAGTCAACAAAAATTCCCTGCAAACAACAGAGGATCAAAAATGGGAACACATGCTCTCAGAATGGAATAAAGAAAAGATCAAACTCATGAATGCAATGATTGGTCCCTCACAAAACTGGCTAGACTTGAAACGCACTCCTGAACCTCCTAGTATTGCTGATGCTCCTAAAAAATTTGGACATTCAATGTTGGATAATGTTGAAGCAGCATATGCCAGACAGGTTCATCAGTATAATAAATATGTATTCCAAGGTGCTAAATCAAGAACAGCTTTGCATCAGAAATTTGCTCAGGTAGCTGAGGACTTCAATGACCCTAAAGTAAAGGAGATGTGGGATATAATAAATACGATGGCCAATATCCCAGCCCTGGTGAGAGATGAAGATCCACTGAAAGCAAGAGGCAATCCAGCAATACAACAGTGTCTTGTCGCACAAGGAAAGAAATACTTACAGAAAAGGTACAAACTATACATGAATGATGTAGTACGTGCCAACCCTGCGGCTGCCCTCCGCGGCGGCGAACCTGGTACATACCCACTTGTTCGAGGATTTGTAGGGCTACGTCTTCAGGGACAGAATTCACAAGGTTTCACTGATGGAGTCATAGATGAACGCCCACTCTGGCCCATGGTTTACTATTGTTTAAGGAGCGGAGATCCAAATGCGGCATTACATTGTTTGAGAAAAGCTGGGCGTGATCATGAAGAATTCATAGCTGCTTTAGAAGAGTATATCCGAAACCCTGAAAAACCACTAAGCGACAAACTGCAGACAGCCATAAATTTCCAATACAGAATACAAGTTCGGAATTCTACAGATCCTTATAAAAGAGCAGTGTACTGTGTGATTGGTTGCTGCGACGTGAGCGACGAGCACTACGAGGTGGCGCGTACGGCCGACGACTACCTCTGGTTGAAACTCTCCTTCATAAGGACGCGCAGTGCCAACGAAACGGAATCGTTCTCTTATTCAGACCTACAGAAACTTATTCTGGAGGAATATGGTGAAACGCATTATCATGCTTATGAAAAACCGGTCGTGTACTTTCAAGTGTTGGCTCTGACTGGACAGTTTGAGCCAGCTATCGAATTCCTCTCGAGAATACCAAGATATCAAGTACACGGTGTTCATATGGCTCTTGCGTTACATGACGTTTACATGCTGGGCACACCGAGGAATGTTCAAGCTCCACTCCTGTCTGTGGACACAGAAGATCCACCACCGTTGCGTCGTTTGAACTTAGCGAGACTGTTACTGCTTTACGTTAGAAAATTTGAACTGACTGATCCCTCTGATGCTTTGCATTACTACTACTTCTTAAGGAATCTAAAAGATCCGAGCGGTAAGAATTTATTCATGTGCTGCTGCACAGACTTGGCATTGGAGAGTAGAGATTACGAGCTCCTATTTGGTCGAATTGACGCTAATTCAAGTTTGAGAAGCCCTGGATTGGTGGATCAATTCAATAACCCTCATATTGACAGCAAGGTGATTGCGTTGAATGTTGCTGAGCAGCTCGTAAACAAGGGCTTATTTGAAGATGCTATTACCATGTACGATATTGCAGATAATTTGGAAAAGGTCGTAGAGTTGTCCTGCGTATTGCTAGCGCAGGTGGTGAATAGCGGCGGCGGGGAAACCGGAGTGCGCGGTCGGCTCGCTAAACGATGTGAACTGACTGACGCGAGGCTACGGGCCTCCACCGAAACCTTACCGCCGTCATTGCTACACGCCTATACCAAGCTCTGCAAACTCTGCATCTTCTTCGATCAGTTCCACGCTGACAATTACGATGCGGCACTTGAGACACTAGGCGAGTGCGAGGTGGTGCCGCTGAACGCGTCGGAGCTGGAGGCGCGCGTGGCGGCGGCGCGGAGCGGGCGCGGGGAGCTGCTGCGCTGCCTGCCCGCCGTGCTGCGCGCGCTGTGCACCGTGCTGCTCGCGCACCGCCAGCGGCTGCGGGCCAACGAGCACTCGCACCCGCTCTCCCAACAGAACAATAAGCAATTGGAGTGGCTTCGTGAACAAGCTGAAGTCCTGAATACTTTTGCTGGCAACATCGCATACAGAATGCCTGGAGACACTTACAGTCAATTAGCGCAAATGCAGATACTTATGCACTGA
Protein
MGDEGFSKLLQQAEQLTTEIEGNEELPRVERSLGQVLEASQELYSRVIQSGANDIQAHLLFGSKGIDLNQISQKLETLSSKRTFEPLQPIADSDIESFLKNEKENAILSLIDEVNKNSLQTTEDQKWEHMLSEWNKEKIKLMNAMIGPSQNWLDLKRTPEPPSIADAPKKFGHSMLDNVEAAYARQVHQYNKYVFQGAKSRTALHQKFAQVAEDFNDPKVKEMWDIINTMANIPALVRDEDPLKARGNPAIQQCLVAQGKKYLQKRYKLYMNDVVRANPAAALRGGEPGTYPLVRGFVGLRLQGQNSQGFTDGVIDERPLWPMVYYCLRSGDPNAALHCLRKAGRDHEEFIAALEEYIRNPEKPLSDKLQTAINFQYRIQVRNSTDPYKRAVYCVIGCCDVSDEHYEVARTADDYLWLKLSFIRTRSANETESFSYSDLQKLILEEYGETHYHAYEKPVVYFQVLALTGQFEPAIEFLSRIPRYQVHGVHMALALHDVYMLGTPRNVQAPLLSVDTEDPPPLRRLNLARLLLLYVRKFELTDPSDALHYYYFLRNLKDPSGKNLFMCCCTDLALESRDYELLFGRIDANSSLRSPGLVDQFNNPHIDSKVIALNVAEQLVNKGLFEDAITMYDIADNLEKVVELSCVLLAQVVNSGGGETGVRGRLAKRCELTDARLRASTETLPPSLLHAYTKLCKLCIFFDQFHADNYDAALETLGECEVVPLNASELEARVAAARSGRGELLRCLPAVLRALCTVLLAHRQRLRANEHSHPLSQQNNKQLEWLREQAEVLNTFAGNIAYRMPGDTYSQLAQMQILMH
Summary
Similarity
Belongs to the nucleoporin interacting component (NIC) family.
Uniprot
H9J0T9
A0A2H1VJB7
A0A2A4JA62
A0A194R408
A0A194PNT3
A0A212FPA1
+ More
S4PXX0 A0A067QXW4 Q16PB8 A0A182QY79 A0A1Q3F3U8 A0A182PN06 B0WEZ2 A0A182NKY1 A0A2J7QBV3 A0A182MLE5 A0A182R843 A0A182VS95 A0A182U4P2 A0A182HJV8 A0A182VJJ8 A0A182KX43 A0A182XK33 Q7QDG3 A0A182K725 A0A182GML5 A0A0K8VFS4 W5JH23 A0A182YM52 A0A1I8MDI7 A0A034W1H3 W8CA93 A0A182F8K1 A0A0A1XP07 A0A2M4AGS5 A0A182JLC6 A0A0A1XAA6 A0A2M4AGK7 A0A1I8Q599 A0A0L0CE31 A0A1A9Y9C1 A0A1B0C4E4 A0A1A9VS81 A0A1B0G339 A0A1A9ZC26 A0A2M4BEF9 A0A1B0DB27 A0A1L8DX32 A0A1W4XKT3 A0A1A9WVB1 D6WKG2 A0A336LSA7 U4UHU5 N6TX90 A0A0T6AX89 A0A1B0CSG9 A0A1A9ZUH8 A0A1B0FQX9 A0A0C9R632 A0A1A9YLY3 A0A158NK52 A0A151X2D5 A0A195BB08 A0A195FBM3 A0A154P729 A0A195EHX1 F4W8D9 A0A1B0BD71 A0A026WRV6 A0A195C2U1 A0A1Y1MEP1 A0A1A9VW89 E9IRE6 A0A0N0BFC6 A0A0J7KXB3 E2A134 K7IMH1 A0A232EG25 A0A310SBT0 A0A088A1B8 A0A1S3KGV4 A0A2A3ELY7 A0A0L7R5H7 E2BTY8 A0A1B6GUF0 A0A0K8SPT8 A0A0P6GRK8 A0A0A9ZIC7 A0A1B6CIL9 E0W256 E9G2T3 A0A0P5YKV3 A0A069DX57 A0A224XJN3 A0A0V0G7D4 T1I6K2 A0A1J1J3H3 A0A2G8JJA3 A0A2C9JNU0 A7S3U8
S4PXX0 A0A067QXW4 Q16PB8 A0A182QY79 A0A1Q3F3U8 A0A182PN06 B0WEZ2 A0A182NKY1 A0A2J7QBV3 A0A182MLE5 A0A182R843 A0A182VS95 A0A182U4P2 A0A182HJV8 A0A182VJJ8 A0A182KX43 A0A182XK33 Q7QDG3 A0A182K725 A0A182GML5 A0A0K8VFS4 W5JH23 A0A182YM52 A0A1I8MDI7 A0A034W1H3 W8CA93 A0A182F8K1 A0A0A1XP07 A0A2M4AGS5 A0A182JLC6 A0A0A1XAA6 A0A2M4AGK7 A0A1I8Q599 A0A0L0CE31 A0A1A9Y9C1 A0A1B0C4E4 A0A1A9VS81 A0A1B0G339 A0A1A9ZC26 A0A2M4BEF9 A0A1B0DB27 A0A1L8DX32 A0A1W4XKT3 A0A1A9WVB1 D6WKG2 A0A336LSA7 U4UHU5 N6TX90 A0A0T6AX89 A0A1B0CSG9 A0A1A9ZUH8 A0A1B0FQX9 A0A0C9R632 A0A1A9YLY3 A0A158NK52 A0A151X2D5 A0A195BB08 A0A195FBM3 A0A154P729 A0A195EHX1 F4W8D9 A0A1B0BD71 A0A026WRV6 A0A195C2U1 A0A1Y1MEP1 A0A1A9VW89 E9IRE6 A0A0N0BFC6 A0A0J7KXB3 E2A134 K7IMH1 A0A232EG25 A0A310SBT0 A0A088A1B8 A0A1S3KGV4 A0A2A3ELY7 A0A0L7R5H7 E2BTY8 A0A1B6GUF0 A0A0K8SPT8 A0A0P6GRK8 A0A0A9ZIC7 A0A1B6CIL9 E0W256 E9G2T3 A0A0P5YKV3 A0A069DX57 A0A224XJN3 A0A0V0G7D4 T1I6K2 A0A1J1J3H3 A0A2G8JJA3 A0A2C9JNU0 A7S3U8
Pubmed
19121390
26354079
22118469
23622113
24845553
17510324
+ More
20966253 12364791 14747013 17210077 26483478 20920257 23761445 25244985 25315136 25348373 24495485 25830018 26108605 18362917 19820115 23537049 21347285 21719571 24508170 30249741 28004739 21282665 20798317 20075255 28648823 25401762 26823975 20566863 21292972 26334808 29023486 15562597 17615350
20966253 12364791 14747013 17210077 26483478 20920257 23761445 25244985 25315136 25348373 24495485 25830018 26108605 18362917 19820115 23537049 21347285 21719571 24508170 30249741 28004739 21282665 20798317 20075255 28648823 25401762 26823975 20566863 21292972 26334808 29023486 15562597 17615350
EMBL
BABH01010335
ODYU01002883
SOQ40923.1
NWSH01002294
PCG68646.1
KQ460779
+ More
KPJ12259.1 KQ459598 KPI94643.1 AGBW02003224 OWR55571.1 GAIX01004303 JAA88257.1 KK853129 KDR10998.1 CH477788 EAT36220.1 AXCN02000107 GFDL01012832 JAV22213.1 DS231912 EDS25850.1 NEVH01016296 PNF26074.1 AXCM01004252 APCN01002209 AAAB01008851 EAA07366.5 JXUM01001746 KQ560127 KXJ84321.1 GDHF01026959 GDHF01014588 JAI25355.1 JAI37726.1 ADMH02001543 ETN62114.1 GAKP01009541 JAC49411.1 GAMC01005966 JAC00590.1 GBXI01001208 JAD13084.1 GGFK01006591 MBW39912.1 GBXI01006627 JAD07665.1 GGFK01006599 MBW39920.1 JRES01000509 KNC30501.1 JXJN01025426 CCAG010020713 GGFJ01002242 MBW51383.1 AJVK01000653 AJVK01000654 GFDF01003064 JAV11020.1 KQ971342 EFA03584.1 UFQT01000137 SSX20705.1 KB632219 ERL90181.1 APGK01051372 APGK01051373 KB741190 ENN73001.1 LJIG01022646 KRT79461.1 AJWK01026051 CCAG010023846 GBYB01003445 JAG73212.1 ADTU01018662 KQ982579 KYQ54561.1 KQ976537 KYM81405.1 KQ981693 KYN37612.1 KQ434819 KZC06930.1 KQ978881 KYN27848.1 GL887908 EGI69430.1 JXJN01012355 KK107119 QOIP01000009 EZA58757.1 RLU18405.1 KQ978344 KYM94910.1 GEZM01033303 JAV84272.1 GL765107 EFZ16893.1 KQ435803 KOX73131.1 LBMM01002347 KMQ94948.1 GL435652 EFN72854.1 NNAY01004881 OXU17301.1 KQ764881 OAD54318.1 KZ288215 PBC32765.1 KQ414652 KOC66137.1 GL450531 EFN80887.1 GECZ01003728 JAS66041.1 GBRD01011004 JAG54820.1 GDIQ01030712 LRGB01000512 JAN64025.1 KZS18548.1 GBHO01000999 GBHO01000998 GDHC01013077 GDHC01009684 JAG42605.1 JAG42606.1 JAQ05552.1 JAQ08945.1 GEDC01024066 JAS13232.1 DS235874 EEB19712.1 GL732530 EFX86453.1 GDIP01057982 JAM45733.1 GBGD01000498 JAC88391.1 GFTR01008083 JAW08343.1 GECL01002168 JAP03956.1 ACPB03019922 CVRI01000066 CRL06014.1 MRZV01001806 PIK35818.1 DS469575 EDO41551.1
KPJ12259.1 KQ459598 KPI94643.1 AGBW02003224 OWR55571.1 GAIX01004303 JAA88257.1 KK853129 KDR10998.1 CH477788 EAT36220.1 AXCN02000107 GFDL01012832 JAV22213.1 DS231912 EDS25850.1 NEVH01016296 PNF26074.1 AXCM01004252 APCN01002209 AAAB01008851 EAA07366.5 JXUM01001746 KQ560127 KXJ84321.1 GDHF01026959 GDHF01014588 JAI25355.1 JAI37726.1 ADMH02001543 ETN62114.1 GAKP01009541 JAC49411.1 GAMC01005966 JAC00590.1 GBXI01001208 JAD13084.1 GGFK01006591 MBW39912.1 GBXI01006627 JAD07665.1 GGFK01006599 MBW39920.1 JRES01000509 KNC30501.1 JXJN01025426 CCAG010020713 GGFJ01002242 MBW51383.1 AJVK01000653 AJVK01000654 GFDF01003064 JAV11020.1 KQ971342 EFA03584.1 UFQT01000137 SSX20705.1 KB632219 ERL90181.1 APGK01051372 APGK01051373 KB741190 ENN73001.1 LJIG01022646 KRT79461.1 AJWK01026051 CCAG010023846 GBYB01003445 JAG73212.1 ADTU01018662 KQ982579 KYQ54561.1 KQ976537 KYM81405.1 KQ981693 KYN37612.1 KQ434819 KZC06930.1 KQ978881 KYN27848.1 GL887908 EGI69430.1 JXJN01012355 KK107119 QOIP01000009 EZA58757.1 RLU18405.1 KQ978344 KYM94910.1 GEZM01033303 JAV84272.1 GL765107 EFZ16893.1 KQ435803 KOX73131.1 LBMM01002347 KMQ94948.1 GL435652 EFN72854.1 NNAY01004881 OXU17301.1 KQ764881 OAD54318.1 KZ288215 PBC32765.1 KQ414652 KOC66137.1 GL450531 EFN80887.1 GECZ01003728 JAS66041.1 GBRD01011004 JAG54820.1 GDIQ01030712 LRGB01000512 JAN64025.1 KZS18548.1 GBHO01000999 GBHO01000998 GDHC01013077 GDHC01009684 JAG42605.1 JAG42606.1 JAQ05552.1 JAQ08945.1 GEDC01024066 JAS13232.1 DS235874 EEB19712.1 GL732530 EFX86453.1 GDIP01057982 JAM45733.1 GBGD01000498 JAC88391.1 GFTR01008083 JAW08343.1 GECL01002168 JAP03956.1 ACPB03019922 CVRI01000066 CRL06014.1 MRZV01001806 PIK35818.1 DS469575 EDO41551.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000027135
+ More
UP000008820 UP000075886 UP000075885 UP000002320 UP000075884 UP000235965 UP000075883 UP000075900 UP000075920 UP000075902 UP000075840 UP000075903 UP000075882 UP000076407 UP000007062 UP000075881 UP000069940 UP000249989 UP000000673 UP000076408 UP000095301 UP000069272 UP000075880 UP000095300 UP000037069 UP000092443 UP000092460 UP000078200 UP000092444 UP000092445 UP000092462 UP000192223 UP000091820 UP000007266 UP000030742 UP000019118 UP000092461 UP000005205 UP000075809 UP000078540 UP000078541 UP000076502 UP000078492 UP000007755 UP000053097 UP000279307 UP000078542 UP000053105 UP000036403 UP000000311 UP000002358 UP000215335 UP000005203 UP000085678 UP000242457 UP000053825 UP000008237 UP000076858 UP000009046 UP000000305 UP000015103 UP000183832 UP000230750 UP000076420 UP000001593
UP000008820 UP000075886 UP000075885 UP000002320 UP000075884 UP000235965 UP000075883 UP000075900 UP000075920 UP000075902 UP000075840 UP000075903 UP000075882 UP000076407 UP000007062 UP000075881 UP000069940 UP000249989 UP000000673 UP000076408 UP000095301 UP000069272 UP000075880 UP000095300 UP000037069 UP000092443 UP000092460 UP000078200 UP000092444 UP000092445 UP000092462 UP000192223 UP000091820 UP000007266 UP000030742 UP000019118 UP000092461 UP000005205 UP000075809 UP000078540 UP000078541 UP000076502 UP000078492 UP000007755 UP000053097 UP000279307 UP000078542 UP000053105 UP000036403 UP000000311 UP000002358 UP000215335 UP000005203 UP000085678 UP000242457 UP000053825 UP000008237 UP000076858 UP000009046 UP000000305 UP000015103 UP000183832 UP000230750 UP000076420 UP000001593
PRIDE
Pfam
PF04097 Nic96
Interpro
IPR007231
Nucleoporin_int_Nup93/Nic96
ProteinModelPortal
H9J0T9
A0A2H1VJB7
A0A2A4JA62
A0A194R408
A0A194PNT3
A0A212FPA1
+ More
S4PXX0 A0A067QXW4 Q16PB8 A0A182QY79 A0A1Q3F3U8 A0A182PN06 B0WEZ2 A0A182NKY1 A0A2J7QBV3 A0A182MLE5 A0A182R843 A0A182VS95 A0A182U4P2 A0A182HJV8 A0A182VJJ8 A0A182KX43 A0A182XK33 Q7QDG3 A0A182K725 A0A182GML5 A0A0K8VFS4 W5JH23 A0A182YM52 A0A1I8MDI7 A0A034W1H3 W8CA93 A0A182F8K1 A0A0A1XP07 A0A2M4AGS5 A0A182JLC6 A0A0A1XAA6 A0A2M4AGK7 A0A1I8Q599 A0A0L0CE31 A0A1A9Y9C1 A0A1B0C4E4 A0A1A9VS81 A0A1B0G339 A0A1A9ZC26 A0A2M4BEF9 A0A1B0DB27 A0A1L8DX32 A0A1W4XKT3 A0A1A9WVB1 D6WKG2 A0A336LSA7 U4UHU5 N6TX90 A0A0T6AX89 A0A1B0CSG9 A0A1A9ZUH8 A0A1B0FQX9 A0A0C9R632 A0A1A9YLY3 A0A158NK52 A0A151X2D5 A0A195BB08 A0A195FBM3 A0A154P729 A0A195EHX1 F4W8D9 A0A1B0BD71 A0A026WRV6 A0A195C2U1 A0A1Y1MEP1 A0A1A9VW89 E9IRE6 A0A0N0BFC6 A0A0J7KXB3 E2A134 K7IMH1 A0A232EG25 A0A310SBT0 A0A088A1B8 A0A1S3KGV4 A0A2A3ELY7 A0A0L7R5H7 E2BTY8 A0A1B6GUF0 A0A0K8SPT8 A0A0P6GRK8 A0A0A9ZIC7 A0A1B6CIL9 E0W256 E9G2T3 A0A0P5YKV3 A0A069DX57 A0A224XJN3 A0A0V0G7D4 T1I6K2 A0A1J1J3H3 A0A2G8JJA3 A0A2C9JNU0 A7S3U8
S4PXX0 A0A067QXW4 Q16PB8 A0A182QY79 A0A1Q3F3U8 A0A182PN06 B0WEZ2 A0A182NKY1 A0A2J7QBV3 A0A182MLE5 A0A182R843 A0A182VS95 A0A182U4P2 A0A182HJV8 A0A182VJJ8 A0A182KX43 A0A182XK33 Q7QDG3 A0A182K725 A0A182GML5 A0A0K8VFS4 W5JH23 A0A182YM52 A0A1I8MDI7 A0A034W1H3 W8CA93 A0A182F8K1 A0A0A1XP07 A0A2M4AGS5 A0A182JLC6 A0A0A1XAA6 A0A2M4AGK7 A0A1I8Q599 A0A0L0CE31 A0A1A9Y9C1 A0A1B0C4E4 A0A1A9VS81 A0A1B0G339 A0A1A9ZC26 A0A2M4BEF9 A0A1B0DB27 A0A1L8DX32 A0A1W4XKT3 A0A1A9WVB1 D6WKG2 A0A336LSA7 U4UHU5 N6TX90 A0A0T6AX89 A0A1B0CSG9 A0A1A9ZUH8 A0A1B0FQX9 A0A0C9R632 A0A1A9YLY3 A0A158NK52 A0A151X2D5 A0A195BB08 A0A195FBM3 A0A154P729 A0A195EHX1 F4W8D9 A0A1B0BD71 A0A026WRV6 A0A195C2U1 A0A1Y1MEP1 A0A1A9VW89 E9IRE6 A0A0N0BFC6 A0A0J7KXB3 E2A134 K7IMH1 A0A232EG25 A0A310SBT0 A0A088A1B8 A0A1S3KGV4 A0A2A3ELY7 A0A0L7R5H7 E2BTY8 A0A1B6GUF0 A0A0K8SPT8 A0A0P6GRK8 A0A0A9ZIC7 A0A1B6CIL9 E0W256 E9G2T3 A0A0P5YKV3 A0A069DX57 A0A224XJN3 A0A0V0G7D4 T1I6K2 A0A1J1J3H3 A0A2G8JJA3 A0A2C9JNU0 A7S3U8
PDB
5IJO
E-value=4.02983e-154,
Score=1400
Ontologies
GO
PANTHER
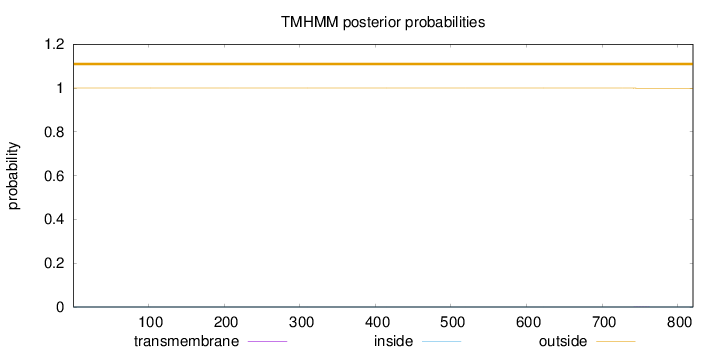
Topology
Subcellular location
Length:
820
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00782
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 820
Population Genetic Test Statistics
Pi
229.962303
Theta
194.024075
Tajima's D
0.579855
CLR
0.089261
CSRT
0.544072796360182
Interpretation
Uncertain
