Gene
KWMTBOMO02197
Pre Gene Modal
BGIBMGA003127
Annotation
PREDICTED:_general_transcription_factor_3C_polypeptide_5_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.923
Sequence
CDS
ATGGAACAGAATAATTTTAGTCATAGTATAGCGGCTGTTCTTTTCCCTGGACTTGTTAAAAACGAAGACAAAGCTATTGAATGTTTGGGTGGTATAAGACAATTGTCTCAGGCTTACACAGAGTTAAACAAAAAACGTCTTGGTCTATCATATCAGCCACAAAATAAACTAGCAAAGAAAATTTATGGCGACTGCGTCAATACTGCTGGTGTGCTCCTGAAAGTTAAAGTAAAAAAAACCATAAATGGCAGTGAAGTGAAAAGGGAGGTAATTTCGACCACCCCACTAGGACAAGTCAGGAAAATATATAAATTTGAATCTTTGTGTGATTTTCAATATTTGCCTGTGAGCAAAGAAGGTGCAGGCTCTGCTCCTCCGCAGTGTATATTAGAACAAATCATGCCTTCTGGAGTTGATACTCTACAATTTTTAACGGAACCAGGACCACAGTTCATAATACCATCAAGTTTCTCGAGATTTGACAAACCTTTAAATCTCGATTATATTGAGAAGAAGAGCATTCTAGAAAAGAACACATCACTTGGTCAAGATGATGATATACACAGAAAAAGAAGACTGGAGCGAGGTGGACAACCAACAGGATTGAAATTCAGCTTAACTGAAGAATTTCCTAAAGAACCACATGAGTACCATTTAAATCAAAAGAGTGAGAGGATGTCTGTTTACCCAGAACTTCAGAGAGATTATGAAGCTGTAAAGAAGATATTTGAAGAAAGGCCGATTTGTTCACAAAATTACATTAAATACCACACTAAAATAAAATTAGTGTTATTAAAAATAATAATGCCATGTTTGGCATTTTATATGATAGATGGTCCATGGAGAACTATGTATATTAAATTTGGATATGACCCTAGAAAGGATCCAAGCTCTCGTAGATACCAAACTTTGGATTTCAGAGTGAGACACCAAGCTGGTTTGAGGTCTATGGTAGTACATTATAAAAAGTCCGAAAGGATACGTAACAAGAGGCGCAAGCAGCCGGATGAGTCCTTTGGAACTGAAGAGAATGTCCCCGAGGCTGTCATATATTTTAGACCTGACACTATGATTTCACAACGACAAGTTTACTATCAGATATGCGATGTACAGCTGCCCGAAGTTCAGGAGATATTAGCTTTGGAGCCTCCAGTTGGATTCGTGTGCCACGAAAAACGTGGCTGGCTTCCTCCGAACACGACCGATGTCTGTCGCGATCACATGTCACGTTATACCAAAGAAGCCCTGCTCTCTAACTTTACTGCTGACATGAAGCTAGAGCACGCTTTAAGTGACGACGACTCCAACACAGATGAAGATATGAATTTAGGCGATACAAATGAAATGGTTCACTAA
Protein
MEQNNFSHSIAAVLFPGLVKNEDKAIECLGGIRQLSQAYTELNKKRLGLSYQPQNKLAKKIYGDCVNTAGVLLKVKVKKTINGSEVKREVISTTPLGQVRKIYKFESLCDFQYLPVSKEGAGSAPPQCILEQIMPSGVDTLQFLTEPGPQFIIPSSFSRFDKPLNLDYIEKKSILEKNTSLGQDDDIHRKRRLERGGQPTGLKFSLTEEFPKEPHEYHLNQKSERMSVYPELQRDYEAVKKIFEERPICSQNYIKYHTKIKLVLLKIIMPCLAFYMIDGPWRTMYIKFGYDPRKDPSSRRYQTLDFRVRHQAGLRSMVVHYKKSERIRNKRRKQPDESFGTEENVPEAVIYFRPDTMISQRQVYYQICDVQLPEVQEILALEPPVGFVCHEKRGWLPPNTTDVCRDHMSRYTKEALLSNFTADMKLEHALSDDDSNTDEDMNLGDTNEMVH
Summary
Uniprot
H9J0U0
A0A194PU33
A0A2A4JB46
A0A194R4U1
A0A2H1VJF0
A0A212FBA8
+ More
E0VNE6 A0A067QXL9 A0A336K515 A0A1B6DG43 A0A1B6FP34 A0A0A9XTA8 A0A1B6J335 Q16W48 A0A224XMU0 A0A1S4FMD1 A0A0P4W0S4 A0A182YNG4 A0A182UUI6 A0A182L6J4 A0A1B0C8R7 A0A1S4JKH6 B0WIR2 A0A182K1S1 A0A1B6LPS5 B4KKG1 A0A1I8MBH0 A0A182G5N9 A0A084VM61 A0A1Q3F4E3 A0A034VQI9 Q7PSE7 A0A1S4H0Q3 A0A0P4W4R1 A0A182VUL0 A0A182Q8M7 A0A2T7NMM8 A0A182RAV4 A0A1A9XGT8 A0A1B0BJF5 A0A1B0B7M4 A0A034VN36 A0A240PMP5 A0A1B0A542 A0A1A9V9J9 A0A0P5GWH2 A0A162T0L4 B4M937 A0A1B0FDE3 B4JPA7 A0A0B6Y834 A0A1W4XK46 T1HR27 A0A293LM76 W8B472 A0A0P5LCP5 A0A0P6GDZ1 A0A0P6DV97 A0A0K8V1Q3 A0A0P5RG53 A0A0P6GE83 A0A0P5SXP4 A0A182SNC7 A0A0P4XF75 A0A158NNB1 A0A2M4BMD3 A0A0M4E7W0 D6WN73 A0A1W4W9G8 A0A182M0C0 K1P552 B4Q9H1 E9FVE5 B4I5S0 A0A182HGC5 A0A131XXW5 A0A147BND5 A0A0P5P463 W5LA77 Q9VIZ2 Q5BIF0 A0A1S3WHI3 B3NM87 A7RMW5 G3IIM7 A1L1K6 A0A023EQ15 A0A1U7RBU5 H0WZE8 A0A3M6T717 A0A1U7R1E3 A0A0P5BF59 A0A0C9PPL3 B7QJM5 T1J199
E0VNE6 A0A067QXL9 A0A336K515 A0A1B6DG43 A0A1B6FP34 A0A0A9XTA8 A0A1B6J335 Q16W48 A0A224XMU0 A0A1S4FMD1 A0A0P4W0S4 A0A182YNG4 A0A182UUI6 A0A182L6J4 A0A1B0C8R7 A0A1S4JKH6 B0WIR2 A0A182K1S1 A0A1B6LPS5 B4KKG1 A0A1I8MBH0 A0A182G5N9 A0A084VM61 A0A1Q3F4E3 A0A034VQI9 Q7PSE7 A0A1S4H0Q3 A0A0P4W4R1 A0A182VUL0 A0A182Q8M7 A0A2T7NMM8 A0A182RAV4 A0A1A9XGT8 A0A1B0BJF5 A0A1B0B7M4 A0A034VN36 A0A240PMP5 A0A1B0A542 A0A1A9V9J9 A0A0P5GWH2 A0A162T0L4 B4M937 A0A1B0FDE3 B4JPA7 A0A0B6Y834 A0A1W4XK46 T1HR27 A0A293LM76 W8B472 A0A0P5LCP5 A0A0P6GDZ1 A0A0P6DV97 A0A0K8V1Q3 A0A0P5RG53 A0A0P6GE83 A0A0P5SXP4 A0A182SNC7 A0A0P4XF75 A0A158NNB1 A0A2M4BMD3 A0A0M4E7W0 D6WN73 A0A1W4W9G8 A0A182M0C0 K1P552 B4Q9H1 E9FVE5 B4I5S0 A0A182HGC5 A0A131XXW5 A0A147BND5 A0A0P5P463 W5LA77 Q9VIZ2 Q5BIF0 A0A1S3WHI3 B3NM87 A7RMW5 G3IIM7 A1L1K6 A0A023EQ15 A0A1U7RBU5 H0WZE8 A0A3M6T717 A0A1U7R1E3 A0A0P5BF59 A0A0C9PPL3 B7QJM5 T1J199
Pubmed
19121390
26354079
22118469
20566863
24845553
25401762
+ More
26823975 17510324 25244985 20966253 17994087 25315136 26483478 24438588 25348373 12364791 27129103 24495485 21347285 18362917 19820115 22992520 22936249 21292972 29652888 25329095 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17615350 21804562 29704459 15489334 15057822 24945155 30382153
26823975 17510324 25244985 20966253 17994087 25315136 26483478 24438588 25348373 12364791 27129103 24495485 21347285 18362917 19820115 22992520 22936249 21292972 29652888 25329095 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17615350 21804562 29704459 15489334 15057822 24945155 30382153
EMBL
BABH01010335
BABH01010336
KQ459598
KPI94645.1
NWSH01002294
PCG68642.1
+ More
KQ460779 KPJ12260.1 ODYU01002883 SOQ40926.1 AGBW02009357 OWR51025.1 AAZO01008544 DS235336 EEB14902.1 KK852840 KDR15195.1 UFQS01000123 UFQT01000123 SSX00066.1 SSX20446.1 GEDC01012644 JAS24654.1 GECZ01017850 JAS51919.1 GBHO01039597 GBHO01020693 GBHO01020685 GBRD01007157 GBRD01007156 GBRD01007155 GDHC01010374 GDHC01004151 GDHC01003154 JAG04007.1 JAG22911.1 JAG22919.1 JAG58664.1 JAQ08255.1 JAQ14478.1 JAQ15475.1 GECU01014107 JAS93599.1 CH477575 EAT38805.1 GFTR01006524 JAW09902.1 GDRN01101449 JAI58371.1 AJWK01001311 DS231951 EDS28639.1 GEBQ01029500 GEBQ01014463 JAT10477.1 JAT25514.1 CH933807 EDW11611.1 JXUM01146429 KQ570172 KXJ68430.1 ATLV01014579 KE524975 KFB39055.1 GFDL01012609 JAV22436.1 GAKP01014253 JAC44699.1 AAAB01008835 EAA05774.5 GDKW01000262 JAI56333.1 AXCN02000844 PZQS01000011 PVD22430.1 JXJN01015380 JXJN01009687 GAKP01014251 JAC44701.1 GDIQ01257918 JAJ93806.1 LRGB01000024 KZS21791.1 CH940654 EDW57713.1 CCAG010008798 CH916372 EDV98737.1 HACG01005454 CEK52319.1 ACPB03024613 GFWV01004239 MAA28969.1 GAMC01014697 JAB91858.1 GDIQ01171414 JAK80311.1 GDIQ01034621 JAN60116.1 GDIQ01086943 JAN07794.1 GDHF01019470 JAI32844.1 GDIQ01101970 JAL49756.1 GDIQ01036161 JAN58576.1 GDIP01140938 JAL62776.1 GDIP01245157 JAI78244.1 ADTU01002654 ADTU01002655 ADTU01002656 ADTU01002657 ADTU01002658 GGFJ01005099 MBW54240.1 CP012525 ALC42866.1 KQ971342 EFA03073.1 AXCM01001945 JH816674 EKC18757.1 CM000361 CM002910 EDX05478.1 KMY90951.1 GL732525 EFX88540.1 CH480822 EDW55726.1 APCN01004213 GEFM01005375 JAP70421.1 GEGO01003101 JAR92303.1 GDIQ01134029 JAL17697.1 AE014134 AAF53767.1 BT021274 AAX33422.1 CH954179 EDV54687.1 DS469521 EDO47172.1 JH003061 RAZU01000235 EGW14225.1 RLQ63974.1 AC129847 BC129109 AAI29110.1 GAPW01001956 JAC11642.1 AAQR03116679 RCHS01004182 RMX37197.1 GDIP01186086 JAJ37316.1 GBYB01003143 JAG72910.1 ABJB010404778 ABJB010545262 ABJB010921981 ABJB011027011 DS953309 EEC19047.1 JH431781
KQ460779 KPJ12260.1 ODYU01002883 SOQ40926.1 AGBW02009357 OWR51025.1 AAZO01008544 DS235336 EEB14902.1 KK852840 KDR15195.1 UFQS01000123 UFQT01000123 SSX00066.1 SSX20446.1 GEDC01012644 JAS24654.1 GECZ01017850 JAS51919.1 GBHO01039597 GBHO01020693 GBHO01020685 GBRD01007157 GBRD01007156 GBRD01007155 GDHC01010374 GDHC01004151 GDHC01003154 JAG04007.1 JAG22911.1 JAG22919.1 JAG58664.1 JAQ08255.1 JAQ14478.1 JAQ15475.1 GECU01014107 JAS93599.1 CH477575 EAT38805.1 GFTR01006524 JAW09902.1 GDRN01101449 JAI58371.1 AJWK01001311 DS231951 EDS28639.1 GEBQ01029500 GEBQ01014463 JAT10477.1 JAT25514.1 CH933807 EDW11611.1 JXUM01146429 KQ570172 KXJ68430.1 ATLV01014579 KE524975 KFB39055.1 GFDL01012609 JAV22436.1 GAKP01014253 JAC44699.1 AAAB01008835 EAA05774.5 GDKW01000262 JAI56333.1 AXCN02000844 PZQS01000011 PVD22430.1 JXJN01015380 JXJN01009687 GAKP01014251 JAC44701.1 GDIQ01257918 JAJ93806.1 LRGB01000024 KZS21791.1 CH940654 EDW57713.1 CCAG010008798 CH916372 EDV98737.1 HACG01005454 CEK52319.1 ACPB03024613 GFWV01004239 MAA28969.1 GAMC01014697 JAB91858.1 GDIQ01171414 JAK80311.1 GDIQ01034621 JAN60116.1 GDIQ01086943 JAN07794.1 GDHF01019470 JAI32844.1 GDIQ01101970 JAL49756.1 GDIQ01036161 JAN58576.1 GDIP01140938 JAL62776.1 GDIP01245157 JAI78244.1 ADTU01002654 ADTU01002655 ADTU01002656 ADTU01002657 ADTU01002658 GGFJ01005099 MBW54240.1 CP012525 ALC42866.1 KQ971342 EFA03073.1 AXCM01001945 JH816674 EKC18757.1 CM000361 CM002910 EDX05478.1 KMY90951.1 GL732525 EFX88540.1 CH480822 EDW55726.1 APCN01004213 GEFM01005375 JAP70421.1 GEGO01003101 JAR92303.1 GDIQ01134029 JAL17697.1 AE014134 AAF53767.1 BT021274 AAX33422.1 CH954179 EDV54687.1 DS469521 EDO47172.1 JH003061 RAZU01000235 EGW14225.1 RLQ63974.1 AC129847 BC129109 AAI29110.1 GAPW01001956 JAC11642.1 AAQR03116679 RCHS01004182 RMX37197.1 GDIP01186086 JAJ37316.1 GBYB01003143 JAG72910.1 ABJB010404778 ABJB010545262 ABJB010921981 ABJB011027011 DS953309 EEC19047.1 JH431781
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000009046
+ More
UP000027135 UP000008820 UP000076408 UP000075903 UP000075882 UP000092461 UP000002320 UP000075881 UP000009192 UP000095301 UP000069940 UP000249989 UP000030765 UP000007062 UP000075920 UP000075886 UP000245119 UP000075900 UP000092443 UP000092460 UP000075880 UP000092445 UP000078200 UP000076858 UP000008792 UP000092444 UP000001070 UP000192223 UP000015103 UP000075901 UP000005205 UP000092553 UP000007266 UP000192221 UP000075883 UP000005408 UP000000304 UP000000305 UP000001292 UP000075840 UP000018467 UP000000803 UP000079721 UP000008711 UP000001593 UP000001075 UP000273346 UP000002494 UP000189706 UP000005225 UP000275408 UP000001555
UP000027135 UP000008820 UP000076408 UP000075903 UP000075882 UP000092461 UP000002320 UP000075881 UP000009192 UP000095301 UP000069940 UP000249989 UP000030765 UP000007062 UP000075920 UP000075886 UP000245119 UP000075900 UP000092443 UP000092460 UP000075880 UP000092445 UP000078200 UP000076858 UP000008792 UP000092444 UP000001070 UP000192223 UP000015103 UP000075901 UP000005205 UP000092553 UP000007266 UP000192221 UP000075883 UP000005408 UP000000304 UP000000305 UP000001292 UP000075840 UP000018467 UP000000803 UP000079721 UP000008711 UP000001593 UP000001075 UP000273346 UP000002494 UP000189706 UP000005225 UP000275408 UP000001555
Pfam
Interpro
IPR041499
Tau95_N
+ More
IPR040454 TF_IIIC_su-5
IPR019136 TF_IIIC_su-5_HTH
IPR001107 Band_7
IPR038407 v-SNARE_N_sf
IPR007705 Vesicle_trsprt_v-SNARE_N
IPR010989 SNARE
IPR000163 Prohibitin
IPR036013 Band_7/SPFH_dom_sf
IPR001936 RasGAP_dom
IPR003123 VPS9
IPR037191 VPS9_dom_sf
IPR041545 DUF5601
IPR008936 Rho_GTPase_activation_prot
IPR040454 TF_IIIC_su-5
IPR019136 TF_IIIC_su-5_HTH
IPR001107 Band_7
IPR038407 v-SNARE_N_sf
IPR007705 Vesicle_trsprt_v-SNARE_N
IPR010989 SNARE
IPR000163 Prohibitin
IPR036013 Band_7/SPFH_dom_sf
IPR001936 RasGAP_dom
IPR003123 VPS9
IPR037191 VPS9_dom_sf
IPR041545 DUF5601
IPR008936 Rho_GTPase_activation_prot
Gene 3D
ProteinModelPortal
H9J0U0
A0A194PU33
A0A2A4JB46
A0A194R4U1
A0A2H1VJF0
A0A212FBA8
+ More
E0VNE6 A0A067QXL9 A0A336K515 A0A1B6DG43 A0A1B6FP34 A0A0A9XTA8 A0A1B6J335 Q16W48 A0A224XMU0 A0A1S4FMD1 A0A0P4W0S4 A0A182YNG4 A0A182UUI6 A0A182L6J4 A0A1B0C8R7 A0A1S4JKH6 B0WIR2 A0A182K1S1 A0A1B6LPS5 B4KKG1 A0A1I8MBH0 A0A182G5N9 A0A084VM61 A0A1Q3F4E3 A0A034VQI9 Q7PSE7 A0A1S4H0Q3 A0A0P4W4R1 A0A182VUL0 A0A182Q8M7 A0A2T7NMM8 A0A182RAV4 A0A1A9XGT8 A0A1B0BJF5 A0A1B0B7M4 A0A034VN36 A0A240PMP5 A0A1B0A542 A0A1A9V9J9 A0A0P5GWH2 A0A162T0L4 B4M937 A0A1B0FDE3 B4JPA7 A0A0B6Y834 A0A1W4XK46 T1HR27 A0A293LM76 W8B472 A0A0P5LCP5 A0A0P6GDZ1 A0A0P6DV97 A0A0K8V1Q3 A0A0P5RG53 A0A0P6GE83 A0A0P5SXP4 A0A182SNC7 A0A0P4XF75 A0A158NNB1 A0A2M4BMD3 A0A0M4E7W0 D6WN73 A0A1W4W9G8 A0A182M0C0 K1P552 B4Q9H1 E9FVE5 B4I5S0 A0A182HGC5 A0A131XXW5 A0A147BND5 A0A0P5P463 W5LA77 Q9VIZ2 Q5BIF0 A0A1S3WHI3 B3NM87 A7RMW5 G3IIM7 A1L1K6 A0A023EQ15 A0A1U7RBU5 H0WZE8 A0A3M6T717 A0A1U7R1E3 A0A0P5BF59 A0A0C9PPL3 B7QJM5 T1J199
E0VNE6 A0A067QXL9 A0A336K515 A0A1B6DG43 A0A1B6FP34 A0A0A9XTA8 A0A1B6J335 Q16W48 A0A224XMU0 A0A1S4FMD1 A0A0P4W0S4 A0A182YNG4 A0A182UUI6 A0A182L6J4 A0A1B0C8R7 A0A1S4JKH6 B0WIR2 A0A182K1S1 A0A1B6LPS5 B4KKG1 A0A1I8MBH0 A0A182G5N9 A0A084VM61 A0A1Q3F4E3 A0A034VQI9 Q7PSE7 A0A1S4H0Q3 A0A0P4W4R1 A0A182VUL0 A0A182Q8M7 A0A2T7NMM8 A0A182RAV4 A0A1A9XGT8 A0A1B0BJF5 A0A1B0B7M4 A0A034VN36 A0A240PMP5 A0A1B0A542 A0A1A9V9J9 A0A0P5GWH2 A0A162T0L4 B4M937 A0A1B0FDE3 B4JPA7 A0A0B6Y834 A0A1W4XK46 T1HR27 A0A293LM76 W8B472 A0A0P5LCP5 A0A0P6GDZ1 A0A0P6DV97 A0A0K8V1Q3 A0A0P5RG53 A0A0P6GE83 A0A0P5SXP4 A0A182SNC7 A0A0P4XF75 A0A158NNB1 A0A2M4BMD3 A0A0M4E7W0 D6WN73 A0A1W4W9G8 A0A182M0C0 K1P552 B4Q9H1 E9FVE5 B4I5S0 A0A182HGC5 A0A131XXW5 A0A147BND5 A0A0P5P463 W5LA77 Q9VIZ2 Q5BIF0 A0A1S3WHI3 B3NM87 A7RMW5 G3IIM7 A1L1K6 A0A023EQ15 A0A1U7RBU5 H0WZE8 A0A3M6T717 A0A1U7R1E3 A0A0P5BF59 A0A0C9PPL3 B7QJM5 T1J199
PDB
4BJI
E-value=6.42351e-11,
Score=163
Ontologies
GO
Topology
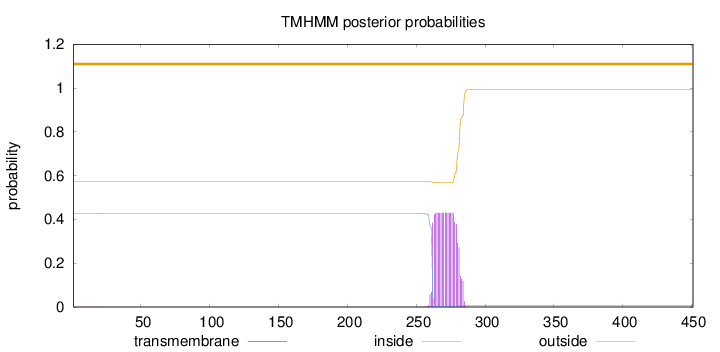
Length:
451
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.72202
Exp number, first 60 AAs:
0.00412
Total prob of N-in:
0.42670
outside
1 - 451
Population Genetic Test Statistics
Pi
23.870214
Theta
21.211305
Tajima's D
1.034361
CLR
0.967429
CSRT
0.670066496675166
Interpretation
Uncertain
