Gene
KWMTBOMO02194
Pre Gene Modal
BGIBMGA003131
Annotation
PREDICTED:_alpha-tocopherol_transfer_protein-like_isoform_X1_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.008 Nuclear Reliability : 1.302
Sequence
CDS
ATGTCAGCTCTTTCGCCGAGAGATCAAAATGGCTGCCGTATTCTAGCGGTAGAATCTGGAGAGAGGTGGAATCCTCGCGAGGTATCTCTGAGGGACATGTTTAGAGTTATACAAATGGGTTTGGTTGGTGCGATATCAGAGCCACGCACACAGATCTGCGGTATTGTATCAATCTTGGATATGAAAGGCTTATCGTTCTCCCACGTCATGCAATTTACTCCATCTTATGCCAAAATGGTCCTGGAGTGGCTTCAGGATTGCGTACCGATTCGTCTTAAGGCTGTTCATATTATAAACCAGCCGTACATATTCAACATGCTATTCGCAATATTCAAGCCGTTTATCAGGGAAAAGCTTCGGGGGCGAATATATCTTCATGGGTCCAATTTGGCGTCTTTACGCGAGCACATTGATCCTGAAGCATTGCGAAAGCGACACGGAGGCTACCTACCAGAGCCAGAAATAGATGGGGAGGTATTATGGAAGATGATGTGTTTTCACGAAAAAGATTTTGAACTGGCCAATTCATACGGATACACAGCGAACAATAATAACAACAAATAA
Protein
MSALSPRDQNGCRILAVESGERWNPREVSLRDMFRVIQMGLVGAISEPRTQICGIVSILDMKGLSFSHVMQFTPSYAKMVLEWLQDCVPIRLKAVHIINQPYIFNMLFAIFKPFIREKLRGRIYLHGSNLASLREHIDPEALRKRHGGYLPEPEIDGEVLWKMMCFHEKDFELANSYGYTANNNNNK
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
H9J0U4
A0A2A4J6E5
A0A194PMV3
A0A194R3J0
A0A2H1VJC8
A0A212FBD1
+ More
A0A2H4RMR1 A0A2H4RMV3 A0A0L7L6B0 Q17LK7 A0A023EP82 A0A182G5R4 W5JLI4 A0A1Q3FZR6 A0A2M4BV85 A0A2M4ANU1 A0A182QS44 A0A182F1U5 W8B3G0 A0A0C9RPS5 A0A182NYZ4 A0A0M4EPM7 A0A182XU66 B3N6K1 A0A182L6N8 Q7QGD5 A0A182IIX2 B4JQP8 B4Q5P8 A0A3B0K8Q6 Q9VM11 B4HY64 A0A034WRP8 A0A0A1WWG2 A0A0K8UIE5 B4NZK5 A0A1W4VWH4 Q29NX9 B4MZV1 B3MKB0 A0A1B0CHK7 B4KKB0 A0A084WV29 B4LS26 A0A1B0D7L7 A0A2P8Y1P9 A0A1I8PS66 A0A1B6EQV2 A0A2H8TCQ8 A0A1L8DYV1 T1PG95 A0A2S2R591 A0A182S5D5 A0A2S2PFK5 C4WVT8 A0A2R7WEM5 A0A182YPC5 E0VDE4 A0A224XTZ4 A0A1I8PHI2 A0A0A1XIQ2 A0A034VGE6 A0A0K8U4J3 A0A0A1X655 A0A034VEC3 A0A1I8MYX3 T1PCI7 A0A0K8V4L7 A0A146LF31 A0A0A9WJF0 A0A182JAJ2 A0A0K8WA35 A0A0C9RMT1 A0A034VH25 A0A0A9WM25 A0A0V0G9I7 A0A0M4EDM6 A0A0A9WRN6 A0A232FIL9 K7J583 A0A0K8SUC5 A0A1A9WKQ8 A0A1B6DJB8 A0A336MDS0 W8B3E2 A0A336MF96 K7J581 W8BDD6 V5GKY7 A0A0A9YIW8 A0A182MCK8 A0A0P4VKI9 E2B177 A0A0P8XGA8 B3MK02 D6WLC7 U5EUE2 B4GJS0 A0A2J7PEI2
A0A2H4RMR1 A0A2H4RMV3 A0A0L7L6B0 Q17LK7 A0A023EP82 A0A182G5R4 W5JLI4 A0A1Q3FZR6 A0A2M4BV85 A0A2M4ANU1 A0A182QS44 A0A182F1U5 W8B3G0 A0A0C9RPS5 A0A182NYZ4 A0A0M4EPM7 A0A182XU66 B3N6K1 A0A182L6N8 Q7QGD5 A0A182IIX2 B4JQP8 B4Q5P8 A0A3B0K8Q6 Q9VM11 B4HY64 A0A034WRP8 A0A0A1WWG2 A0A0K8UIE5 B4NZK5 A0A1W4VWH4 Q29NX9 B4MZV1 B3MKB0 A0A1B0CHK7 B4KKB0 A0A084WV29 B4LS26 A0A1B0D7L7 A0A2P8Y1P9 A0A1I8PS66 A0A1B6EQV2 A0A2H8TCQ8 A0A1L8DYV1 T1PG95 A0A2S2R591 A0A182S5D5 A0A2S2PFK5 C4WVT8 A0A2R7WEM5 A0A182YPC5 E0VDE4 A0A224XTZ4 A0A1I8PHI2 A0A0A1XIQ2 A0A034VGE6 A0A0K8U4J3 A0A0A1X655 A0A034VEC3 A0A1I8MYX3 T1PCI7 A0A0K8V4L7 A0A146LF31 A0A0A9WJF0 A0A182JAJ2 A0A0K8WA35 A0A0C9RMT1 A0A034VH25 A0A0A9WM25 A0A0V0G9I7 A0A0M4EDM6 A0A0A9WRN6 A0A232FIL9 K7J583 A0A0K8SUC5 A0A1A9WKQ8 A0A1B6DJB8 A0A336MDS0 W8B3E2 A0A336MF96 K7J581 W8BDD6 V5GKY7 A0A0A9YIW8 A0A182MCK8 A0A0P4VKI9 E2B177 A0A0P8XGA8 B3MK02 D6WLC7 U5EUE2 B4GJS0 A0A2J7PEI2
Pubmed
19121390
26354079
22118469
29136137
26227816
17510324
+ More
24945155 26483478 20920257 23761445 24495485 17994087 18057021 20966253 12364791 14747013 17210077 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25830018 17550304 15632085 24438588 29403074 25315136 25244985 20566863 26823975 25401762 28648823 20075255 27129103 20798317 18362917 19820115
24945155 26483478 20920257 23761445 24495485 17994087 18057021 20966253 12364791 14747013 17210077 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25830018 17550304 15632085 24438588 29403074 25315136 25244985 20566863 26823975 25401762 28648823 20075255 27129103 20798317 18362917 19820115
EMBL
BABH01010340
NWSH01003077
PCG66992.1
KQ459598
KPI94647.1
KQ460779
+ More
KPJ12262.1 ODYU01002883 SOQ40928.1 AGBW02009357 OWR51027.1 MG434616 ATY51922.1 MG434664 ATY51970.1 JTDY01002730 KOB70834.1 CH477214 EAT47606.1 GAPW01002808 JAC10790.1 JXUM01043917 JXUM01043918 JXUM01043919 JXUM01043920 KQ561381 KXJ78764.1 ADMH02001192 ETN63770.1 GFDL01002053 JAV32992.1 GGFJ01007859 MBW57000.1 GGFK01009128 MBW42449.1 AXCN02000846 GAMC01013468 JAB93087.1 GBYB01009181 JAG78948.1 CP012523 ALC38825.1 CH954177 EDV59217.1 KQS70691.1 AAAB01008835 EAA05796.4 APCN01004203 CH916372 EDV99228.1 CM000361 CM002910 EDX04094.1 KMY88768.1 OUUW01000006 SPP82006.1 AE014134 AY129433 AAF52517.2 AAM76175.1 CH480818 EDW51994.1 GAKP01001935 JAC57017.1 GBXI01013530 GBXI01011090 JAD00762.1 JAD03202.1 GDHF01025842 JAI26472.1 CM000157 EDW87754.1 KRJ97379.1 KRJ97380.1 CH379058 EAL34514.1 CH963920 EDW77886.1 CH902620 EDV32494.1 AJWK01012444 AJWK01012445 CH933807 EDW12641.1 KRG03346.1 ATLV01027276 KL646861 KFB54073.1 CH940649 EDW64712.1 AJVK01027007 PYGN01001044 PSN38173.1 GECZ01029481 GECZ01002739 JAS40288.1 JAS67030.1 GFXV01000075 MBW11880.1 GFDF01002433 JAV11651.1 KA647709 AFP62338.1 GGMS01015921 MBY85124.1 GGMR01015612 MBY28231.1 ABLF02028917 AK341667 BAH72008.1 KK854712 PTY18113.1 DS235073 EEB11400.1 GFTR01004853 JAW11573.1 GBXI01003894 JAD10398.1 GAKP01018105 GAKP01018102 JAC40850.1 GDHF01030836 JAI21478.1 GBXI01008129 JAD06163.1 GAKP01018103 JAC40849.1 KA645663 KA649316 AFP60292.1 GDHF01018508 JAI33806.1 GDHC01011746 JAQ06883.1 GBHO01036068 GBHO01036067 GBRD01009010 JAG07536.1 JAG07537.1 JAG56811.1 AXCP01009230 GDHF01004415 GDHF01004329 JAI47899.1 JAI47985.1 GBYB01014737 JAG84504.1 GAKP01018106 GAKP01018104 JAC40848.1 GBHO01036066 GBRD01009011 GDHC01012350 JAG07538.1 JAG56810.1 JAQ06279.1 GECL01001454 JAP04670.1 ALC39972.1 GBHO01036069 JAG07535.1 NNAY01000143 OXU30604.1 AAZX01006217 GBRD01009012 JAG56809.1 GEDC01011502 JAS25796.1 UFQT01000934 SSX28070.1 GAMC01018760 GAMC01018759 JAB87796.1 SSX28071.1 AAZX01008416 GAMC01018761 JAB87794.1 GALX01003707 JAB64759.1 GBHO01010577 GBRD01013833 GDHC01016075 JAG33027.1 JAG51993.1 JAQ02554.1 AXCM01001490 GDKW01001749 JAI54846.1 GL444799 EFN60569.1 KPU73951.1 EDV32457.1 KPU73950.1 KQ971343 EFA03469.2 GANO01003920 JAB55951.1 CH479184 EDW36886.1 NEVH01026096 PNF14735.1
KPJ12262.1 ODYU01002883 SOQ40928.1 AGBW02009357 OWR51027.1 MG434616 ATY51922.1 MG434664 ATY51970.1 JTDY01002730 KOB70834.1 CH477214 EAT47606.1 GAPW01002808 JAC10790.1 JXUM01043917 JXUM01043918 JXUM01043919 JXUM01043920 KQ561381 KXJ78764.1 ADMH02001192 ETN63770.1 GFDL01002053 JAV32992.1 GGFJ01007859 MBW57000.1 GGFK01009128 MBW42449.1 AXCN02000846 GAMC01013468 JAB93087.1 GBYB01009181 JAG78948.1 CP012523 ALC38825.1 CH954177 EDV59217.1 KQS70691.1 AAAB01008835 EAA05796.4 APCN01004203 CH916372 EDV99228.1 CM000361 CM002910 EDX04094.1 KMY88768.1 OUUW01000006 SPP82006.1 AE014134 AY129433 AAF52517.2 AAM76175.1 CH480818 EDW51994.1 GAKP01001935 JAC57017.1 GBXI01013530 GBXI01011090 JAD00762.1 JAD03202.1 GDHF01025842 JAI26472.1 CM000157 EDW87754.1 KRJ97379.1 KRJ97380.1 CH379058 EAL34514.1 CH963920 EDW77886.1 CH902620 EDV32494.1 AJWK01012444 AJWK01012445 CH933807 EDW12641.1 KRG03346.1 ATLV01027276 KL646861 KFB54073.1 CH940649 EDW64712.1 AJVK01027007 PYGN01001044 PSN38173.1 GECZ01029481 GECZ01002739 JAS40288.1 JAS67030.1 GFXV01000075 MBW11880.1 GFDF01002433 JAV11651.1 KA647709 AFP62338.1 GGMS01015921 MBY85124.1 GGMR01015612 MBY28231.1 ABLF02028917 AK341667 BAH72008.1 KK854712 PTY18113.1 DS235073 EEB11400.1 GFTR01004853 JAW11573.1 GBXI01003894 JAD10398.1 GAKP01018105 GAKP01018102 JAC40850.1 GDHF01030836 JAI21478.1 GBXI01008129 JAD06163.1 GAKP01018103 JAC40849.1 KA645663 KA649316 AFP60292.1 GDHF01018508 JAI33806.1 GDHC01011746 JAQ06883.1 GBHO01036068 GBHO01036067 GBRD01009010 JAG07536.1 JAG07537.1 JAG56811.1 AXCP01009230 GDHF01004415 GDHF01004329 JAI47899.1 JAI47985.1 GBYB01014737 JAG84504.1 GAKP01018106 GAKP01018104 JAC40848.1 GBHO01036066 GBRD01009011 GDHC01012350 JAG07538.1 JAG56810.1 JAQ06279.1 GECL01001454 JAP04670.1 ALC39972.1 GBHO01036069 JAG07535.1 NNAY01000143 OXU30604.1 AAZX01006217 GBRD01009012 JAG56809.1 GEDC01011502 JAS25796.1 UFQT01000934 SSX28070.1 GAMC01018760 GAMC01018759 JAB87796.1 SSX28071.1 AAZX01008416 GAMC01018761 JAB87794.1 GALX01003707 JAB64759.1 GBHO01010577 GBRD01013833 GDHC01016075 JAG33027.1 JAG51993.1 JAQ02554.1 AXCM01001490 GDKW01001749 JAI54846.1 GL444799 EFN60569.1 KPU73951.1 EDV32457.1 KPU73950.1 KQ971343 EFA03469.2 GANO01003920 JAB55951.1 CH479184 EDW36886.1 NEVH01026096 PNF14735.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000008820 UP000069940 UP000249989 UP000000673 UP000075886 UP000069272 UP000075884 UP000092553 UP000076407 UP000008711 UP000075882 UP000007062 UP000075840 UP000001070 UP000000304 UP000268350 UP000000803 UP000001292 UP000002282 UP000192221 UP000001819 UP000007798 UP000007801 UP000092461 UP000009192 UP000030765 UP000008792 UP000092462 UP000245037 UP000095300 UP000095301 UP000075900 UP000007819 UP000076408 UP000009046 UP000075880 UP000215335 UP000002358 UP000091820 UP000075883 UP000000311 UP000007266 UP000008744 UP000235965
UP000008820 UP000069940 UP000249989 UP000000673 UP000075886 UP000069272 UP000075884 UP000092553 UP000076407 UP000008711 UP000075882 UP000007062 UP000075840 UP000001070 UP000000304 UP000268350 UP000000803 UP000001292 UP000002282 UP000192221 UP000001819 UP000007798 UP000007801 UP000092461 UP000009192 UP000030765 UP000008792 UP000092462 UP000245037 UP000095300 UP000095301 UP000075900 UP000007819 UP000076408 UP000009046 UP000075880 UP000215335 UP000002358 UP000091820 UP000075883 UP000000311 UP000007266 UP000008744 UP000235965
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J0U4
A0A2A4J6E5
A0A194PMV3
A0A194R3J0
A0A2H1VJC8
A0A212FBD1
+ More
A0A2H4RMR1 A0A2H4RMV3 A0A0L7L6B0 Q17LK7 A0A023EP82 A0A182G5R4 W5JLI4 A0A1Q3FZR6 A0A2M4BV85 A0A2M4ANU1 A0A182QS44 A0A182F1U5 W8B3G0 A0A0C9RPS5 A0A182NYZ4 A0A0M4EPM7 A0A182XU66 B3N6K1 A0A182L6N8 Q7QGD5 A0A182IIX2 B4JQP8 B4Q5P8 A0A3B0K8Q6 Q9VM11 B4HY64 A0A034WRP8 A0A0A1WWG2 A0A0K8UIE5 B4NZK5 A0A1W4VWH4 Q29NX9 B4MZV1 B3MKB0 A0A1B0CHK7 B4KKB0 A0A084WV29 B4LS26 A0A1B0D7L7 A0A2P8Y1P9 A0A1I8PS66 A0A1B6EQV2 A0A2H8TCQ8 A0A1L8DYV1 T1PG95 A0A2S2R591 A0A182S5D5 A0A2S2PFK5 C4WVT8 A0A2R7WEM5 A0A182YPC5 E0VDE4 A0A224XTZ4 A0A1I8PHI2 A0A0A1XIQ2 A0A034VGE6 A0A0K8U4J3 A0A0A1X655 A0A034VEC3 A0A1I8MYX3 T1PCI7 A0A0K8V4L7 A0A146LF31 A0A0A9WJF0 A0A182JAJ2 A0A0K8WA35 A0A0C9RMT1 A0A034VH25 A0A0A9WM25 A0A0V0G9I7 A0A0M4EDM6 A0A0A9WRN6 A0A232FIL9 K7J583 A0A0K8SUC5 A0A1A9WKQ8 A0A1B6DJB8 A0A336MDS0 W8B3E2 A0A336MF96 K7J581 W8BDD6 V5GKY7 A0A0A9YIW8 A0A182MCK8 A0A0P4VKI9 E2B177 A0A0P8XGA8 B3MK02 D6WLC7 U5EUE2 B4GJS0 A0A2J7PEI2
A0A2H4RMR1 A0A2H4RMV3 A0A0L7L6B0 Q17LK7 A0A023EP82 A0A182G5R4 W5JLI4 A0A1Q3FZR6 A0A2M4BV85 A0A2M4ANU1 A0A182QS44 A0A182F1U5 W8B3G0 A0A0C9RPS5 A0A182NYZ4 A0A0M4EPM7 A0A182XU66 B3N6K1 A0A182L6N8 Q7QGD5 A0A182IIX2 B4JQP8 B4Q5P8 A0A3B0K8Q6 Q9VM11 B4HY64 A0A034WRP8 A0A0A1WWG2 A0A0K8UIE5 B4NZK5 A0A1W4VWH4 Q29NX9 B4MZV1 B3MKB0 A0A1B0CHK7 B4KKB0 A0A084WV29 B4LS26 A0A1B0D7L7 A0A2P8Y1P9 A0A1I8PS66 A0A1B6EQV2 A0A2H8TCQ8 A0A1L8DYV1 T1PG95 A0A2S2R591 A0A182S5D5 A0A2S2PFK5 C4WVT8 A0A2R7WEM5 A0A182YPC5 E0VDE4 A0A224XTZ4 A0A1I8PHI2 A0A0A1XIQ2 A0A034VGE6 A0A0K8U4J3 A0A0A1X655 A0A034VEC3 A0A1I8MYX3 T1PCI7 A0A0K8V4L7 A0A146LF31 A0A0A9WJF0 A0A182JAJ2 A0A0K8WA35 A0A0C9RMT1 A0A034VH25 A0A0A9WM25 A0A0V0G9I7 A0A0M4EDM6 A0A0A9WRN6 A0A232FIL9 K7J583 A0A0K8SUC5 A0A1A9WKQ8 A0A1B6DJB8 A0A336MDS0 W8B3E2 A0A336MF96 K7J581 W8BDD6 V5GKY7 A0A0A9YIW8 A0A182MCK8 A0A0P4VKI9 E2B177 A0A0P8XGA8 B3MK02 D6WLC7 U5EUE2 B4GJS0 A0A2J7PEI2
PDB
3W68
E-value=2.50422e-23,
Score=265
Ontologies
GO
Topology
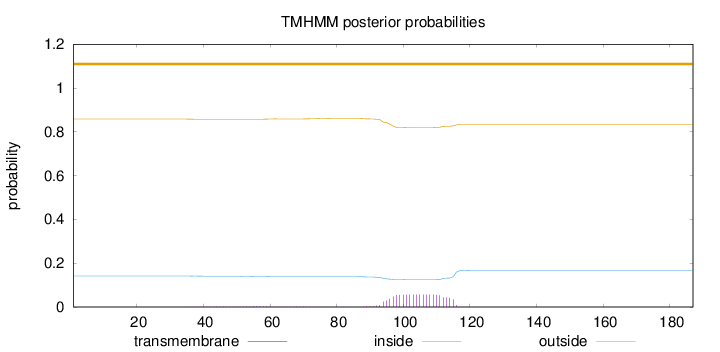
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.20952
Exp number, first 60 AAs:
0.0557
Total prob of N-in:
0.14190
outside
1 - 187
Population Genetic Test Statistics
Pi
230.819445
Theta
191.149008
Tajima's D
0.775204
CLR
245.909873
CSRT
0.593970301484926
Interpretation
Uncertain
