Pre Gene Modal
BGIBMGA003032
Annotation
CRAL-TRIO_domain-containing_protein?_partial_[Manduca_sexta]
Location in the cell
Cytoplasmic Reliability : 2.835
Sequence
CDS
ATGGTGAAATTATCACAACACATGCTGACTGAGTTGGAAAAGCTGCCGGGCGTCCAACTCGGCGACTTCTTACTGCAGTTCGAGTTGGATGAACCACGTGAGTCGGTCAGAGAGATTGCCAGAAGAGAACTTCGAGAAATACCAGAAATCGTCGAACCAGCTGTTGAAGAGTTAAAAAGACTTCTTAGTGAAGATACGGATATGCATGTTCCTCTCAACAATGAAGCTTGGCTAATTAGATTCTTAAGGCCGTGCAAGTTTTATCCGGAGAGCGCCTATGATTTGATAAAGCGTTACTATGGTTTCAAAGTGAAACATCACAAACATTACGATGGCCTAACACCGAGCAAGGAGACTAATGTGTTCAACGAGAACGTGCTGACGGTGCTGCCGACCAGAGATCAGTGCGGGCGGAGGGTGCTCGTCTTGGAACTTGGAAAGAAATGGAAGCATAATAAATGTTCGCTGGACGAGGTTTTCAAAGGCTGCGTGTTGTTCCTTGAGGCGGCCATGTTGGAACCGGAGTCGCAGATCTGCGGAGCTGTCGTCATCTTTGACATGGACGGTCTCTCCATGCAGCAAGTCATGCAGTTCACGCCACCTTTTGCTAAGAGAATCGTAGACTGGTTGCAGGAATGCATACCGCTCCGCATAAAAGGGTTGTACATCATCAACCAGCCGTACGTCTTCAATATGGTGTTCCAGCTTTTCAAGCCATTCCTAAAGGAGAAGCTTCGGTCGCGCATCATCTTCATGGGCAACGATCGCGAGCTGTTACACAAACACATCAGCCCCAAGTGTCTGCCGGACTGCTATGGAGGAACGCTCACAATACCAAAAGTGACAGGCCCTCAGTGGCTGGAACTACTCCTCATGTGCGATCAAGAGTTTATTGCAATTAATTCGTTCGGCTACAAAAAGAAATGA
Protein
MVKLSQHMLTELEKLPGVQLGDFLLQFELDEPRESVREIARRELREIPEIVEPAVEELKRLLSEDTDMHVPLNNEAWLIRFLRPCKFYPESAYDLIKRYYGFKVKHHKHYDGLTPSKETNVFNENVLTVLPTRDQCGRRVLVLELGKKWKHNKCSLDEVFKGCVLFLEAAMLEPESQICGAVVIFDMDGLSMQQVMQFTPPFAKRIVDWLQECIPLRIKGLYIINQPYVFNMVFQLFKPFLKEKLRSRIIFMGNDRELLHKHISPKCLPDCYGGTLTIPKVTGPQWLELLLMCDQEFIAINSFGYKKK
Summary
Uniprot
H9J0J5
A0A0S2Z361
A0A1E1WJQ6
A0A1E1WLD7
A0A2A4J6E8
A0A194R380
+ More
A0A194PNT8 A0A2H1VJB8 A0A2H4RMQ5 A0A2H4RMU4 A0A034VGE6 A0A034VH25 A0A0C9RMT1 A0A034VEC3 A0A0K8WA35 A0A0K8U4J3 A0A0K8V4L7 A0A0A1XIQ2 A0A0A1X655 W8B3E2 W8BDD6 E0VDE5 B4KK66 V5GKY7 A0A0Q9X452 A0A0Q9XE97 B4JP83 D6WLC6 A0A0Q9VYU2 A0A232FIL9 K7J583 B4M965 A0A0Q9VYM6 B4MYQ2 A0A0P8XGA8 A0A0Q9WR39 B3MK02 A0A0P8XGT8 A0A0M4EDM6 A0A310SPW5 A0A2J7PEI2 V9IE24 A0A154PQQ1 Q29K50 B4GWW5 A0A1B6FQ38 A0A1B6GEL9 A0A088A1C5 A0A0R3NUW9 A0A0R3NVA0 A0A0L7QVL5 A0A151XFR1 A0A1W4VF84 A0A3B0JTQ6 A0A1W4V2H6 A0A1W4VER3 A0A3B0KI99 A0A3B0JTM8 A0A3B0JWM1 A0A3B0JTP7 A0A1Y1N505 A0A2P8Y1P9 A0A1B6CSP3 A0A195E222 A0A1B6CR84 A0A1B6CQL7 Q9I7M3 A0A0R1DVB0 A0A0R1DPF2 Q8INW5 A0A195FW06 A0A0Q5VZ23 J3JV36 A0A0Q5VW94 B3NM56 Q9VIW5 A0A0J9R4K2 A0A0J9R425 B4PAY5 C6TP94 A0A1B0CF11 B4Q9Y0 N6TAY2 A0A0J7KZ52 B4I5V1 E2C3S5 A0A195D4G6 E9J2R6 A0A2M4BUQ6 A0A2M4BV92 A0A0T6B1L9 A0A2M4BSV4 A0A1S4GY60 B0WGK3 Q7PNC9 A0A1Q3FMW6 A0A182RCQ9 A0A182UN81 A0A182UFG0 A0A182L1B3
A0A194PNT8 A0A2H1VJB8 A0A2H4RMQ5 A0A2H4RMU4 A0A034VGE6 A0A034VH25 A0A0C9RMT1 A0A034VEC3 A0A0K8WA35 A0A0K8U4J3 A0A0K8V4L7 A0A0A1XIQ2 A0A0A1X655 W8B3E2 W8BDD6 E0VDE5 B4KK66 V5GKY7 A0A0Q9X452 A0A0Q9XE97 B4JP83 D6WLC6 A0A0Q9VYU2 A0A232FIL9 K7J583 B4M965 A0A0Q9VYM6 B4MYQ2 A0A0P8XGA8 A0A0Q9WR39 B3MK02 A0A0P8XGT8 A0A0M4EDM6 A0A310SPW5 A0A2J7PEI2 V9IE24 A0A154PQQ1 Q29K50 B4GWW5 A0A1B6FQ38 A0A1B6GEL9 A0A088A1C5 A0A0R3NUW9 A0A0R3NVA0 A0A0L7QVL5 A0A151XFR1 A0A1W4VF84 A0A3B0JTQ6 A0A1W4V2H6 A0A1W4VER3 A0A3B0KI99 A0A3B0JTM8 A0A3B0JWM1 A0A3B0JTP7 A0A1Y1N505 A0A2P8Y1P9 A0A1B6CSP3 A0A195E222 A0A1B6CR84 A0A1B6CQL7 Q9I7M3 A0A0R1DVB0 A0A0R1DPF2 Q8INW5 A0A195FW06 A0A0Q5VZ23 J3JV36 A0A0Q5VW94 B3NM56 Q9VIW5 A0A0J9R4K2 A0A0J9R425 B4PAY5 C6TP94 A0A1B0CF11 B4Q9Y0 N6TAY2 A0A0J7KZ52 B4I5V1 E2C3S5 A0A195D4G6 E9J2R6 A0A2M4BUQ6 A0A2M4BV92 A0A0T6B1L9 A0A2M4BSV4 A0A1S4GY60 B0WGK3 Q7PNC9 A0A1Q3FMW6 A0A182RCQ9 A0A182UN81 A0A182UFG0 A0A182L1B3
Pubmed
19121390
25684408
26354079
29136137
25348373
25830018
+ More
24495485 20566863 17994087 18362917 19820115 28648823 20075255 18057021 15632085 28004739 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22516182 23537049 22936249 20798317 21282665 12364791 20966253
24495485 20566863 17994087 18362917 19820115 28648823 20075255 18057021 15632085 28004739 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22516182 23537049 22936249 20798317 21282665 12364791 20966253
EMBL
BABH01010342
KT943564
ALQ33322.1
GDQN01003928
JAT87126.1
GDQN01006564
+ More
GDQN01003388 JAT84490.1 JAT87666.1 NWSH01003077 PCG66993.1 KQ460779 KPJ12263.1 KQ459598 KPI94648.1 ODYU01002883 SOQ40929.1 MG434614 ATY51920.1 MG434655 ATY51961.1 GAKP01018105 GAKP01018102 JAC40850.1 GAKP01018106 GAKP01018104 JAC40848.1 GBYB01014737 JAG84504.1 GAKP01018103 JAC40849.1 GDHF01004415 GDHF01004329 JAI47899.1 JAI47985.1 GDHF01030836 JAI21478.1 GDHF01018508 JAI33806.1 GBXI01003894 JAD10398.1 GBXI01008129 JAD06163.1 GAMC01018760 GAMC01018759 JAB87796.1 GAMC01018761 JAB87794.1 DS235073 EEB11401.1 CH933807 EDW11584.1 GALX01003707 JAB64759.1 KRG02783.1 KRG02782.1 CH916372 EDV98713.1 KQ971343 EFA04094.2 CH940654 KRF77928.1 NNAY01000143 OXU30604.1 AAZX01006217 EDW57741.1 KRF77929.1 CH963913 EDW77241.2 CH902620 KPU73951.1 KRF98609.1 EDV32457.1 KPU73950.1 KPU73952.1 CP012523 ALC39972.1 KQ760343 OAD60850.1 NEVH01026096 PNF14735.1 JR039441 AEY58706.1 KQ435050 KZC14222.1 CH379061 EAL33328.2 CH479195 EDW27292.1 GECZ01017444 JAS52325.1 GECZ01008877 JAS60892.1 KRT04904.1 KRT04905.1 KQ414725 KOC62672.1 KQ982194 KYQ59108.1 OUUW01000010 SPP85485.1 SPP85486.1 SPP85487.1 SPP85483.1 SPP85484.1 GEZM01015302 JAV91765.1 PYGN01001044 PSN38173.1 GEDC01020729 JAS16569.1 KQ979763 KYN19195.1 GEDC01021336 JAS15962.1 GEDC01021634 JAS15664.1 AE014134 AAG22441.1 AFH03765.1 CM000158 KRJ99168.1 KRJ99167.1 BT125736 AAN11041.1 ADP21875.1 AGB93112.1 KQ981208 KYN44840.1 CH954179 KQS61887.1 BT127102 KB632364 AEE62064.1 ERL93557.1 KQS61886.1 EDV54656.1 AY061463 AAF53799.2 AAL29011.1 CM002910 KMY91013.1 KMY91011.1 KMY91012.1 EDW90414.1 BT099580 ACU43542.1 AJWK01009541 CM000361 EDX05510.1 KMY91010.1 APGK01037684 KB740948 ENN77439.1 LBMM01001723 KMQ95842.1 CH480822 EDW55757.1 GL452364 EFN77311.1 KQ976870 KYN07795.1 GL767998 EFZ12890.1 GGFJ01007602 MBW56743.1 GGFJ01007537 MBW56678.1 LJIG01016210 KRT81286.1 GGFJ01006951 MBW56092.1 AAAB01008964 DS231927 EDS27032.1 EAA12394.4 GFDL01006116 JAV28929.1
GDQN01003388 JAT84490.1 JAT87666.1 NWSH01003077 PCG66993.1 KQ460779 KPJ12263.1 KQ459598 KPI94648.1 ODYU01002883 SOQ40929.1 MG434614 ATY51920.1 MG434655 ATY51961.1 GAKP01018105 GAKP01018102 JAC40850.1 GAKP01018106 GAKP01018104 JAC40848.1 GBYB01014737 JAG84504.1 GAKP01018103 JAC40849.1 GDHF01004415 GDHF01004329 JAI47899.1 JAI47985.1 GDHF01030836 JAI21478.1 GDHF01018508 JAI33806.1 GBXI01003894 JAD10398.1 GBXI01008129 JAD06163.1 GAMC01018760 GAMC01018759 JAB87796.1 GAMC01018761 JAB87794.1 DS235073 EEB11401.1 CH933807 EDW11584.1 GALX01003707 JAB64759.1 KRG02783.1 KRG02782.1 CH916372 EDV98713.1 KQ971343 EFA04094.2 CH940654 KRF77928.1 NNAY01000143 OXU30604.1 AAZX01006217 EDW57741.1 KRF77929.1 CH963913 EDW77241.2 CH902620 KPU73951.1 KRF98609.1 EDV32457.1 KPU73950.1 KPU73952.1 CP012523 ALC39972.1 KQ760343 OAD60850.1 NEVH01026096 PNF14735.1 JR039441 AEY58706.1 KQ435050 KZC14222.1 CH379061 EAL33328.2 CH479195 EDW27292.1 GECZ01017444 JAS52325.1 GECZ01008877 JAS60892.1 KRT04904.1 KRT04905.1 KQ414725 KOC62672.1 KQ982194 KYQ59108.1 OUUW01000010 SPP85485.1 SPP85486.1 SPP85487.1 SPP85483.1 SPP85484.1 GEZM01015302 JAV91765.1 PYGN01001044 PSN38173.1 GEDC01020729 JAS16569.1 KQ979763 KYN19195.1 GEDC01021336 JAS15962.1 GEDC01021634 JAS15664.1 AE014134 AAG22441.1 AFH03765.1 CM000158 KRJ99168.1 KRJ99167.1 BT125736 AAN11041.1 ADP21875.1 AGB93112.1 KQ981208 KYN44840.1 CH954179 KQS61887.1 BT127102 KB632364 AEE62064.1 ERL93557.1 KQS61886.1 EDV54656.1 AY061463 AAF53799.2 AAL29011.1 CM002910 KMY91013.1 KMY91011.1 KMY91012.1 EDW90414.1 BT099580 ACU43542.1 AJWK01009541 CM000361 EDX05510.1 KMY91010.1 APGK01037684 KB740948 ENN77439.1 LBMM01001723 KMQ95842.1 CH480822 EDW55757.1 GL452364 EFN77311.1 KQ976870 KYN07795.1 GL767998 EFZ12890.1 GGFJ01007602 MBW56743.1 GGFJ01007537 MBW56678.1 LJIG01016210 KRT81286.1 GGFJ01006951 MBW56092.1 AAAB01008964 DS231927 EDS27032.1 EAA12394.4 GFDL01006116 JAV28929.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000009046
UP000009192
+ More
UP000001070 UP000007266 UP000008792 UP000215335 UP000002358 UP000007798 UP000007801 UP000092553 UP000235965 UP000076502 UP000001819 UP000008744 UP000005203 UP000053825 UP000075809 UP000192221 UP000268350 UP000245037 UP000078492 UP000000803 UP000002282 UP000078541 UP000008711 UP000030742 UP000092461 UP000000304 UP000019118 UP000036403 UP000001292 UP000008237 UP000078542 UP000002320 UP000007062 UP000075900 UP000075903 UP000075902 UP000075882
UP000001070 UP000007266 UP000008792 UP000215335 UP000002358 UP000007798 UP000007801 UP000092553 UP000235965 UP000076502 UP000001819 UP000008744 UP000005203 UP000053825 UP000075809 UP000192221 UP000268350 UP000245037 UP000078492 UP000000803 UP000002282 UP000078541 UP000008711 UP000030742 UP000092461 UP000000304 UP000019118 UP000036403 UP000001292 UP000008237 UP000078542 UP000002320 UP000007062 UP000075900 UP000075903 UP000075902 UP000075882
Pfam
PF00650 CRAL_TRIO
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J0J5
A0A0S2Z361
A0A1E1WJQ6
A0A1E1WLD7
A0A2A4J6E8
A0A194R380
+ More
A0A194PNT8 A0A2H1VJB8 A0A2H4RMQ5 A0A2H4RMU4 A0A034VGE6 A0A034VH25 A0A0C9RMT1 A0A034VEC3 A0A0K8WA35 A0A0K8U4J3 A0A0K8V4L7 A0A0A1XIQ2 A0A0A1X655 W8B3E2 W8BDD6 E0VDE5 B4KK66 V5GKY7 A0A0Q9X452 A0A0Q9XE97 B4JP83 D6WLC6 A0A0Q9VYU2 A0A232FIL9 K7J583 B4M965 A0A0Q9VYM6 B4MYQ2 A0A0P8XGA8 A0A0Q9WR39 B3MK02 A0A0P8XGT8 A0A0M4EDM6 A0A310SPW5 A0A2J7PEI2 V9IE24 A0A154PQQ1 Q29K50 B4GWW5 A0A1B6FQ38 A0A1B6GEL9 A0A088A1C5 A0A0R3NUW9 A0A0R3NVA0 A0A0L7QVL5 A0A151XFR1 A0A1W4VF84 A0A3B0JTQ6 A0A1W4V2H6 A0A1W4VER3 A0A3B0KI99 A0A3B0JTM8 A0A3B0JWM1 A0A3B0JTP7 A0A1Y1N505 A0A2P8Y1P9 A0A1B6CSP3 A0A195E222 A0A1B6CR84 A0A1B6CQL7 Q9I7M3 A0A0R1DVB0 A0A0R1DPF2 Q8INW5 A0A195FW06 A0A0Q5VZ23 J3JV36 A0A0Q5VW94 B3NM56 Q9VIW5 A0A0J9R4K2 A0A0J9R425 B4PAY5 C6TP94 A0A1B0CF11 B4Q9Y0 N6TAY2 A0A0J7KZ52 B4I5V1 E2C3S5 A0A195D4G6 E9J2R6 A0A2M4BUQ6 A0A2M4BV92 A0A0T6B1L9 A0A2M4BSV4 A0A1S4GY60 B0WGK3 Q7PNC9 A0A1Q3FMW6 A0A182RCQ9 A0A182UN81 A0A182UFG0 A0A182L1B3
A0A194PNT8 A0A2H1VJB8 A0A2H4RMQ5 A0A2H4RMU4 A0A034VGE6 A0A034VH25 A0A0C9RMT1 A0A034VEC3 A0A0K8WA35 A0A0K8U4J3 A0A0K8V4L7 A0A0A1XIQ2 A0A0A1X655 W8B3E2 W8BDD6 E0VDE5 B4KK66 V5GKY7 A0A0Q9X452 A0A0Q9XE97 B4JP83 D6WLC6 A0A0Q9VYU2 A0A232FIL9 K7J583 B4M965 A0A0Q9VYM6 B4MYQ2 A0A0P8XGA8 A0A0Q9WR39 B3MK02 A0A0P8XGT8 A0A0M4EDM6 A0A310SPW5 A0A2J7PEI2 V9IE24 A0A154PQQ1 Q29K50 B4GWW5 A0A1B6FQ38 A0A1B6GEL9 A0A088A1C5 A0A0R3NUW9 A0A0R3NVA0 A0A0L7QVL5 A0A151XFR1 A0A1W4VF84 A0A3B0JTQ6 A0A1W4V2H6 A0A1W4VER3 A0A3B0KI99 A0A3B0JTM8 A0A3B0JWM1 A0A3B0JTP7 A0A1Y1N505 A0A2P8Y1P9 A0A1B6CSP3 A0A195E222 A0A1B6CR84 A0A1B6CQL7 Q9I7M3 A0A0R1DVB0 A0A0R1DPF2 Q8INW5 A0A195FW06 A0A0Q5VZ23 J3JV36 A0A0Q5VW94 B3NM56 Q9VIW5 A0A0J9R4K2 A0A0J9R425 B4PAY5 C6TP94 A0A1B0CF11 B4Q9Y0 N6TAY2 A0A0J7KZ52 B4I5V1 E2C3S5 A0A195D4G6 E9J2R6 A0A2M4BUQ6 A0A2M4BV92 A0A0T6B1L9 A0A2M4BSV4 A0A1S4GY60 B0WGK3 Q7PNC9 A0A1Q3FMW6 A0A182RCQ9 A0A182UN81 A0A182UFG0 A0A182L1B3
PDB
3HY5
E-value=3.97428e-31,
Score=335
Ontologies
GO
Topology
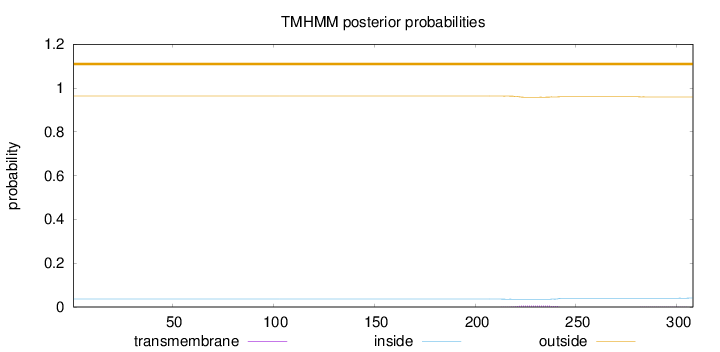
Length:
308
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15236
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03663
outside
1 - 308
Population Genetic Test Statistics
Pi
209.222594
Theta
165.498967
Tajima's D
0.633521
CLR
0.989535
CSRT
0.550622468876556
Interpretation
Uncertain
