Gene
KWMTBOMO02192
Pre Gene Modal
BGIBMGA003132
Annotation
CRAL-TRIO_domain-containing_protein?_partial_[Manduca_sexta]
Location in the cell
Cytoplasmic Reliability : 3.074
Sequence
CDS
ATGCCGACTCCGGATTACAGCGTGGACTTGGACCTCGGCGAGCCCCCGCCAGAGATCCAAGACTACGCTCGTCAGCATTGCGGCGAGGACCCCAATACTCGACTTCAAGCCATCTACGAACTAAGGGACATGATTTATGAGCGCGGTGAATGTACCCCTCACAGGATGGATGATGAATTCCTGATTAGATTCCTTCGTGCTCGAAATTTCATACCACAAAAAGCTCATAGATTGCTCGTAAACTACTATCAGTTCAGAGCAGAGAATCCGGATTTGTTCGAGAACATTCATCCTATGGATCTTCACGCCATCGGGGACGCAGATATCATGACGGTACCACCGTACCGCGGACCCGATGGGCGCAGACTCATTATATATAAATTTGGTAATTGGGACCCAAAGACCATTTGCGTGGAAGAATTGTTCAAAGCAACAATCCTTAGCCTAGAACTGGGTATGATGGAGCCGCGTACTCAGATCCTGGGGGCAAGTGCTCTGGTCGACCTGGAGGATCTTGGCTCCCAGCATGTGTGGCAGTGTACGCCGTCTGTAGCAGCCAAATTACTCAAGATACTTGTGAATTGCTACCCAACGAAGATACACGCGATCCACATAATCAATCACTCCTGGATCTTCGACAAAGCTTACAGCGTTTTTAAGCCATTCTTGAACGACGTCATGAAATCGAGGATCTATTTCCACGGTAATGACGTCACATCGCTTCACAAGCACATCAGCCCGGACTATTTACCCGAAAGCTATGGCGGTGTCTGGCCGGATTACTCCTACACTATTTGGTTGGATTCTTTGCGTAAAAACGTGGTCGTCGCCAAGGAAATGATATCATGTGGTTACAAATTCCGTGAACATGAATTAAACCAAGACGTCCTTGAACATTTGACTAAGGAAGGCATTCAAATATCCTAA
Protein
MPTPDYSVDLDLGEPPPEIQDYARQHCGEDPNTRLQAIYELRDMIYERGECTPHRMDDEFLIRFLRARNFIPQKAHRLLVNYYQFRAENPDLFENIHPMDLHAIGDADIMTVPPYRGPDGRRLIIYKFGNWDPKTICVEELFKATILSLELGMMEPRTQILGASALVDLEDLGSQHVWQCTPSVAAKLLKILVNCYPTKIHAIHIINHSWIFDKAYSVFKPFLNDVMKSRIYFHGNDVTSLHKHISPDYLPESYGGVWPDYSYTIWLDSLRKNVVVAKEMISCGYKFREHELNQDVLEHLTKEGIQIS
Summary
Uniprot
H9J0U5
A0A0S2Z2X8
A0A2H4RMR4
S4PDY3
A0A2H4RMU9
A0A2A4J639
+ More
A0A194PPJ0 A0A212FFX7 A0A0L7LMG0 A0A194R4U3 D6WK77 A0A2J7PEI5 A0A151XFF6 A0A0L7QVN0 A0A154PSY4 A0A195D4M7 A0A158NHN1 A0A2H1VTH5 A0A088A1P4 K7J584 A0A195AZZ6 A0A0M8ZXJ1 A0A2P8Y1N2 A0A088AQZ8 A0A026WVT7 A0A195EF31 A0A3L8DGB1 E2B175 A0A195FWL3 A0A1Y1MAD0 E2C3S6 F4WD80 A0A0C9RS57 A0A1Y1M6Q3 E9J2R5 J3JWX5 A0A067R7A4 A0A1Y1KBD3 A0A1Y1KBD8 A0A310SSY3 A0A069DR99 U5EUI3 B0WIP9 A0A1S4JIR2 A0A1Q3FJS7 A0A1L8DYZ0 N6THZ4 R4G2V6 A0A1B6CWG4 A0A224XT63 A0A1L8DYX8 U4UM51 A0A023F8M2 W5JFN1 A0A336N2P7 A0A171B1L9 A0A2M3ZAW7 A0A1B6EH81 T1DG07 A0A023ENW0 A0A2M4BVI7 Q16T91 A0A2M4BVL7 A0A182USL8 A0A182M7C8 A0A2C9H8Q1 A0A182U9P9 A0A2M4ADQ0 A0A2M4ADP7 A0A182KCU4 A0A1S4H0R7 A0A182GRY0 A0A182JEP7 A0A1B6IJ80 A0A1J1HZC9 A0A182SLW3 A0A084VM70 A0A2S2QBB3 A0A034WVU7 A0A182QDV4 A0A3B0KI89 A0A3B0JTP5 A0A3B0JTL6 A0A2H8TKU5 J9JPQ9 A0A0R3NV55 B4N164 Q29K54 A0A0K8VQT4 A0A0R3P0C1 A0A2S2PKI9 B4GWX2 A0A1W4V301 A0A1W4VFB1 A0A0A9VYR6 A0A182P9H0 A0A0K8SWM7 B3NM52 B4Q9Y4 Q9VIW1
A0A194PPJ0 A0A212FFX7 A0A0L7LMG0 A0A194R4U3 D6WK77 A0A2J7PEI5 A0A151XFF6 A0A0L7QVN0 A0A154PSY4 A0A195D4M7 A0A158NHN1 A0A2H1VTH5 A0A088A1P4 K7J584 A0A195AZZ6 A0A0M8ZXJ1 A0A2P8Y1N2 A0A088AQZ8 A0A026WVT7 A0A195EF31 A0A3L8DGB1 E2B175 A0A195FWL3 A0A1Y1MAD0 E2C3S6 F4WD80 A0A0C9RS57 A0A1Y1M6Q3 E9J2R5 J3JWX5 A0A067R7A4 A0A1Y1KBD3 A0A1Y1KBD8 A0A310SSY3 A0A069DR99 U5EUI3 B0WIP9 A0A1S4JIR2 A0A1Q3FJS7 A0A1L8DYZ0 N6THZ4 R4G2V6 A0A1B6CWG4 A0A224XT63 A0A1L8DYX8 U4UM51 A0A023F8M2 W5JFN1 A0A336N2P7 A0A171B1L9 A0A2M3ZAW7 A0A1B6EH81 T1DG07 A0A023ENW0 A0A2M4BVI7 Q16T91 A0A2M4BVL7 A0A182USL8 A0A182M7C8 A0A2C9H8Q1 A0A182U9P9 A0A2M4ADQ0 A0A2M4ADP7 A0A182KCU4 A0A1S4H0R7 A0A182GRY0 A0A182JEP7 A0A1B6IJ80 A0A1J1HZC9 A0A182SLW3 A0A084VM70 A0A2S2QBB3 A0A034WVU7 A0A182QDV4 A0A3B0KI89 A0A3B0JTP5 A0A3B0JTL6 A0A2H8TKU5 J9JPQ9 A0A0R3NV55 B4N164 Q29K54 A0A0K8VQT4 A0A0R3P0C1 A0A2S2PKI9 B4GWX2 A0A1W4V301 A0A1W4VFB1 A0A0A9VYR6 A0A182P9H0 A0A0K8SWM7 B3NM52 B4Q9Y4 Q9VIW1
Pubmed
19121390
25684408
29136137
23622113
26354079
22118469
+ More
26227816 18362917 19820115 21347285 20075255 29403074 24508170 30249741 20798317 28004739 21719571 21282665 22516182 24845553 26334808 23537049 25474469 20920257 23761445 24945155 17510324 12364791 26483478 24438588 25348373 15632085 17994087 25401762 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
26227816 18362917 19820115 21347285 20075255 29403074 24508170 30249741 20798317 28004739 21719571 21282665 22516182 24845553 26334808 23537049 25474469 20920257 23761445 24945155 17510324 12364791 26483478 24438588 25348373 15632085 17994087 25401762 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01010352
BABH01010353
KT943556
ALQ33314.1
MG434618
ATY51924.1
+ More
GAIX01004762 JAA87798.1 MG434649 ATY51955.1 NWSH01003077 PCG66994.1 KQ459598 KPI94649.1 AGBW02008751 OWR52636.1 JTDY01000576 KOB76610.1 KQ460779 KPJ12265.1 KQ971342 EFA03949.1 NEVH01026096 PNF14739.1 KQ982194 KYQ59109.1 KQ414725 KOC62670.1 KQ435050 KZC14220.1 KQ976870 KYN07796.1 ADTU01015717 ODYU01004059 SOQ43544.1 AAZX01000350 KQ976694 KYM77615.1 KQ435803 KOX73150.1 PYGN01001044 PSN38167.1 KK107109 EZA59219.1 KQ979039 KYN23459.1 QOIP01000008 RLU19232.1 GL444799 EFN60567.1 KQ981208 KYN44838.1 GEZM01038761 JAV81520.1 GL452364 EFN77312.1 GL888086 EGI67807.1 GBYB01011455 JAG81222.1 GEZM01038762 JAV81519.1 GL767998 EFZ12889.1 BT127743 AEE62705.1 KK852692 KDR18336.1 GEZM01087248 JAV58822.1 GEZM01087247 JAV58823.1 KQ760343 OAD60852.1 GBGD01002291 JAC86598.1 GANO01002311 JAB57560.1 DS231951 EDS28626.1 GFDL01007164 JAV27881.1 GFDF01002447 JAV11637.1 APGK01037684 KB740948 ENN77438.1 ACPB03010458 GAHY01002256 JAA75254.1 GEDC01019442 JAS17856.1 GFTR01004796 JAW11630.1 GFDF01002448 JAV11636.1 KB632364 ERL93558.1 GBBI01001005 JAC17707.1 ADMH02001356 ETN62876.1 UFQS01002102 UFQT01001089 UFQT01002102 SSX13240.1 SSX28859.1 SSX32678.1 GEMB01000384 JAS02737.1 GGFM01004847 MBW25598.1 GECZ01032471 JAS37298.1 GAMD01002946 JAA98644.1 GAPW01002969 GAPW01002968 JAC10630.1 GGFJ01007959 MBW57100.1 CH477657 EAT37699.1 GGFJ01007958 MBW57099.1 AXCM01005439 GGFK01005595 MBW38916.1 GGFK01005593 MBW38914.1 AAAB01008835 JXUM01083834 KQ563399 KXJ73895.1 GECU01020731 JAS86975.1 CVRI01000037 CRK93463.1 ATLV01014581 KE524975 KFB39064.1 GGMS01005825 MBY75028.1 GAKP01000248 JAC58704.1 AXCN02000844 OUUW01000010 SPP85476.1 SPP85475.1 SPP85477.1 GFXV01002951 MBW14756.1 ABLF02039888 CH379061 KRT04900.1 CH963920 EDW78046.1 EAL33324.2 GDHF01011087 GDHF01007080 JAI41227.1 JAI45234.1 KRT04901.1 GGMR01017285 MBY29904.1 CH479195 EDW27299.1 GBHO01042855 JAG00749.1 GBRD01008212 GBRD01008211 JAG57609.1 CH954179 EDV54652.1 CM000361 CM002910 EDX05514.1 KMY91018.1 AE014134 AY113277 AAF53803.1 AAM29282.1 AAN11044.1
GAIX01004762 JAA87798.1 MG434649 ATY51955.1 NWSH01003077 PCG66994.1 KQ459598 KPI94649.1 AGBW02008751 OWR52636.1 JTDY01000576 KOB76610.1 KQ460779 KPJ12265.1 KQ971342 EFA03949.1 NEVH01026096 PNF14739.1 KQ982194 KYQ59109.1 KQ414725 KOC62670.1 KQ435050 KZC14220.1 KQ976870 KYN07796.1 ADTU01015717 ODYU01004059 SOQ43544.1 AAZX01000350 KQ976694 KYM77615.1 KQ435803 KOX73150.1 PYGN01001044 PSN38167.1 KK107109 EZA59219.1 KQ979039 KYN23459.1 QOIP01000008 RLU19232.1 GL444799 EFN60567.1 KQ981208 KYN44838.1 GEZM01038761 JAV81520.1 GL452364 EFN77312.1 GL888086 EGI67807.1 GBYB01011455 JAG81222.1 GEZM01038762 JAV81519.1 GL767998 EFZ12889.1 BT127743 AEE62705.1 KK852692 KDR18336.1 GEZM01087248 JAV58822.1 GEZM01087247 JAV58823.1 KQ760343 OAD60852.1 GBGD01002291 JAC86598.1 GANO01002311 JAB57560.1 DS231951 EDS28626.1 GFDL01007164 JAV27881.1 GFDF01002447 JAV11637.1 APGK01037684 KB740948 ENN77438.1 ACPB03010458 GAHY01002256 JAA75254.1 GEDC01019442 JAS17856.1 GFTR01004796 JAW11630.1 GFDF01002448 JAV11636.1 KB632364 ERL93558.1 GBBI01001005 JAC17707.1 ADMH02001356 ETN62876.1 UFQS01002102 UFQT01001089 UFQT01002102 SSX13240.1 SSX28859.1 SSX32678.1 GEMB01000384 JAS02737.1 GGFM01004847 MBW25598.1 GECZ01032471 JAS37298.1 GAMD01002946 JAA98644.1 GAPW01002969 GAPW01002968 JAC10630.1 GGFJ01007959 MBW57100.1 CH477657 EAT37699.1 GGFJ01007958 MBW57099.1 AXCM01005439 GGFK01005595 MBW38916.1 GGFK01005593 MBW38914.1 AAAB01008835 JXUM01083834 KQ563399 KXJ73895.1 GECU01020731 JAS86975.1 CVRI01000037 CRK93463.1 ATLV01014581 KE524975 KFB39064.1 GGMS01005825 MBY75028.1 GAKP01000248 JAC58704.1 AXCN02000844 OUUW01000010 SPP85476.1 SPP85475.1 SPP85477.1 GFXV01002951 MBW14756.1 ABLF02039888 CH379061 KRT04900.1 CH963920 EDW78046.1 EAL33324.2 GDHF01011087 GDHF01007080 JAI41227.1 JAI45234.1 KRT04901.1 GGMR01017285 MBY29904.1 CH479195 EDW27299.1 GBHO01042855 JAG00749.1 GBRD01008212 GBRD01008211 JAG57609.1 CH954179 EDV54652.1 CM000361 CM002910 EDX05514.1 KMY91018.1 AE014134 AY113277 AAF53803.1 AAM29282.1 AAN11044.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000053240
+ More
UP000007266 UP000235965 UP000075809 UP000053825 UP000076502 UP000078542 UP000005205 UP000005203 UP000002358 UP000078540 UP000053105 UP000245037 UP000053097 UP000078492 UP000279307 UP000000311 UP000078541 UP000008237 UP000007755 UP000027135 UP000002320 UP000019118 UP000015103 UP000030742 UP000000673 UP000008820 UP000075903 UP000075883 UP000076407 UP000075902 UP000075881 UP000069940 UP000249989 UP000075880 UP000183832 UP000075901 UP000030765 UP000075886 UP000268350 UP000007819 UP000001819 UP000007798 UP000008744 UP000192221 UP000075885 UP000008711 UP000000304 UP000000803
UP000007266 UP000235965 UP000075809 UP000053825 UP000076502 UP000078542 UP000005205 UP000005203 UP000002358 UP000078540 UP000053105 UP000245037 UP000053097 UP000078492 UP000279307 UP000000311 UP000078541 UP000008237 UP000007755 UP000027135 UP000002320 UP000019118 UP000015103 UP000030742 UP000000673 UP000008820 UP000075903 UP000075883 UP000076407 UP000075902 UP000075881 UP000069940 UP000249989 UP000075880 UP000183832 UP000075901 UP000030765 UP000075886 UP000268350 UP000007819 UP000001819 UP000007798 UP000008744 UP000192221 UP000075885 UP000008711 UP000000304 UP000000803
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J0U5
A0A0S2Z2X8
A0A2H4RMR4
S4PDY3
A0A2H4RMU9
A0A2A4J639
+ More
A0A194PPJ0 A0A212FFX7 A0A0L7LMG0 A0A194R4U3 D6WK77 A0A2J7PEI5 A0A151XFF6 A0A0L7QVN0 A0A154PSY4 A0A195D4M7 A0A158NHN1 A0A2H1VTH5 A0A088A1P4 K7J584 A0A195AZZ6 A0A0M8ZXJ1 A0A2P8Y1N2 A0A088AQZ8 A0A026WVT7 A0A195EF31 A0A3L8DGB1 E2B175 A0A195FWL3 A0A1Y1MAD0 E2C3S6 F4WD80 A0A0C9RS57 A0A1Y1M6Q3 E9J2R5 J3JWX5 A0A067R7A4 A0A1Y1KBD3 A0A1Y1KBD8 A0A310SSY3 A0A069DR99 U5EUI3 B0WIP9 A0A1S4JIR2 A0A1Q3FJS7 A0A1L8DYZ0 N6THZ4 R4G2V6 A0A1B6CWG4 A0A224XT63 A0A1L8DYX8 U4UM51 A0A023F8M2 W5JFN1 A0A336N2P7 A0A171B1L9 A0A2M3ZAW7 A0A1B6EH81 T1DG07 A0A023ENW0 A0A2M4BVI7 Q16T91 A0A2M4BVL7 A0A182USL8 A0A182M7C8 A0A2C9H8Q1 A0A182U9P9 A0A2M4ADQ0 A0A2M4ADP7 A0A182KCU4 A0A1S4H0R7 A0A182GRY0 A0A182JEP7 A0A1B6IJ80 A0A1J1HZC9 A0A182SLW3 A0A084VM70 A0A2S2QBB3 A0A034WVU7 A0A182QDV4 A0A3B0KI89 A0A3B0JTP5 A0A3B0JTL6 A0A2H8TKU5 J9JPQ9 A0A0R3NV55 B4N164 Q29K54 A0A0K8VQT4 A0A0R3P0C1 A0A2S2PKI9 B4GWX2 A0A1W4V301 A0A1W4VFB1 A0A0A9VYR6 A0A182P9H0 A0A0K8SWM7 B3NM52 B4Q9Y4 Q9VIW1
A0A194PPJ0 A0A212FFX7 A0A0L7LMG0 A0A194R4U3 D6WK77 A0A2J7PEI5 A0A151XFF6 A0A0L7QVN0 A0A154PSY4 A0A195D4M7 A0A158NHN1 A0A2H1VTH5 A0A088A1P4 K7J584 A0A195AZZ6 A0A0M8ZXJ1 A0A2P8Y1N2 A0A088AQZ8 A0A026WVT7 A0A195EF31 A0A3L8DGB1 E2B175 A0A195FWL3 A0A1Y1MAD0 E2C3S6 F4WD80 A0A0C9RS57 A0A1Y1M6Q3 E9J2R5 J3JWX5 A0A067R7A4 A0A1Y1KBD3 A0A1Y1KBD8 A0A310SSY3 A0A069DR99 U5EUI3 B0WIP9 A0A1S4JIR2 A0A1Q3FJS7 A0A1L8DYZ0 N6THZ4 R4G2V6 A0A1B6CWG4 A0A224XT63 A0A1L8DYX8 U4UM51 A0A023F8M2 W5JFN1 A0A336N2P7 A0A171B1L9 A0A2M3ZAW7 A0A1B6EH81 T1DG07 A0A023ENW0 A0A2M4BVI7 Q16T91 A0A2M4BVL7 A0A182USL8 A0A182M7C8 A0A2C9H8Q1 A0A182U9P9 A0A2M4ADQ0 A0A2M4ADP7 A0A182KCU4 A0A1S4H0R7 A0A182GRY0 A0A182JEP7 A0A1B6IJ80 A0A1J1HZC9 A0A182SLW3 A0A084VM70 A0A2S2QBB3 A0A034WVU7 A0A182QDV4 A0A3B0KI89 A0A3B0JTP5 A0A3B0JTL6 A0A2H8TKU5 J9JPQ9 A0A0R3NV55 B4N164 Q29K54 A0A0K8VQT4 A0A0R3P0C1 A0A2S2PKI9 B4GWX2 A0A1W4V301 A0A1W4VFB1 A0A0A9VYR6 A0A182P9H0 A0A0K8SWM7 B3NM52 B4Q9Y4 Q9VIW1
PDB
3W68
E-value=2.79211e-40,
Score=414
Ontologies
GO
Topology
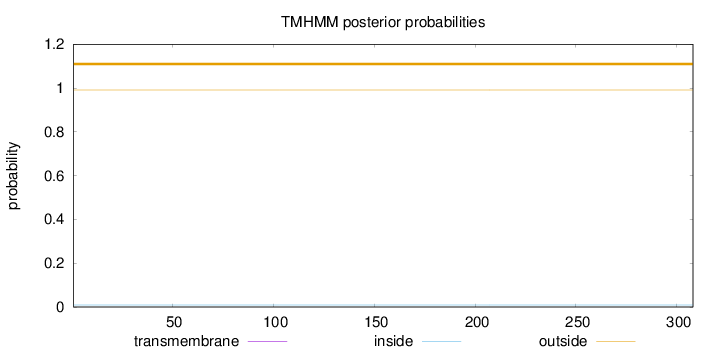
Length:
308
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00236
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00933
outside
1 - 308
Population Genetic Test Statistics
Pi
203.834469
Theta
163.66137
Tajima's D
0.826589
CLR
0.101695
CSRT
0.609469526523674
Interpretation
Uncertain
